|
|
|
|
|
| Sample: |
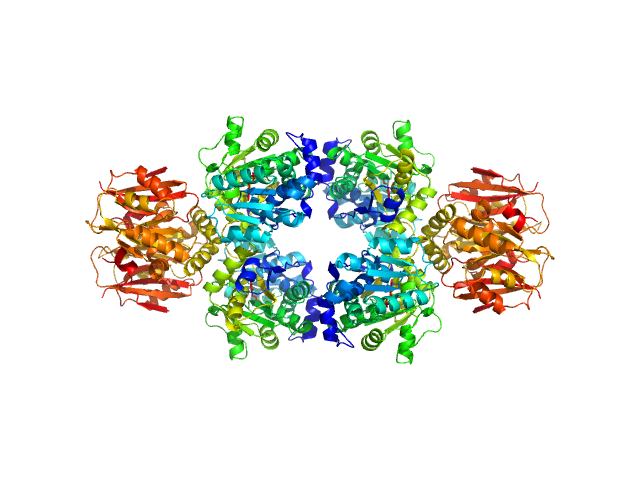
IgA protease (E540A) monomer, 128 kDa Thomasclavelia ramosa protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 May 23
|
Thomasclavelia ramosa IgA peptidase (IgAse) active site mutant (A31-I1166)
Juan Sebastián Ramírez Larrota
|
| RgGuinier |
5.7 |
nm |
| Dmax |
27.9 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
L-threonine dehydratase biosynthetic IlvA tetramer, 233 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
284 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
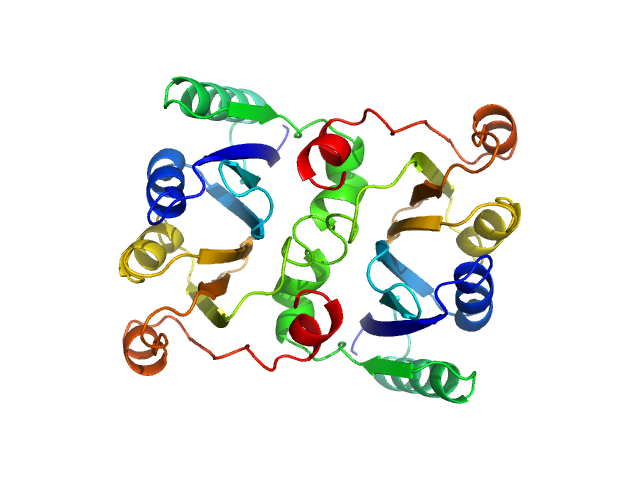
L-threonine dehydratase biosynthetic IlvA Regulatory domain dimer, 46 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
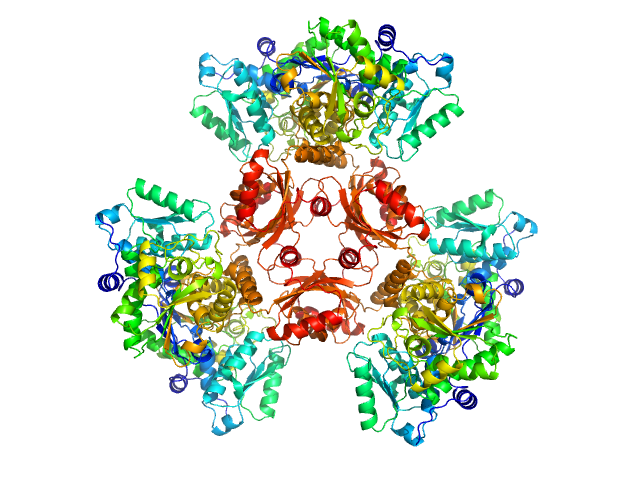
L-threonine dehydratase biosynthetic IlvA hexamer, 288 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
386 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform 2 of B-cell CLL/lymphoma 7 protein family member A monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.8 |
nm |
| Dmax |
28.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
3.3 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein dimer, 55 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 110 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
5.2 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 110 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
5.4 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jan 29
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|