|
|
|
|
|
| Sample: |
Lysyne-specific Demethylase LSD2 monomer, 89 kDa Homo sapiens protein
NPAC linker+DH (delta-205) tetramer, 150 kDa Homo sapiens protein
Histone H3 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 22 kDa Xenopus laevis protein
Histone H2a dimer, 28 kDa Xenopus laevis protein
Histone H2b dimer, 27 kDa Xenopus laevis protein
147bp 601 Widom sequence monomer, 45 kDa synthetic construct DNA
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 11
|
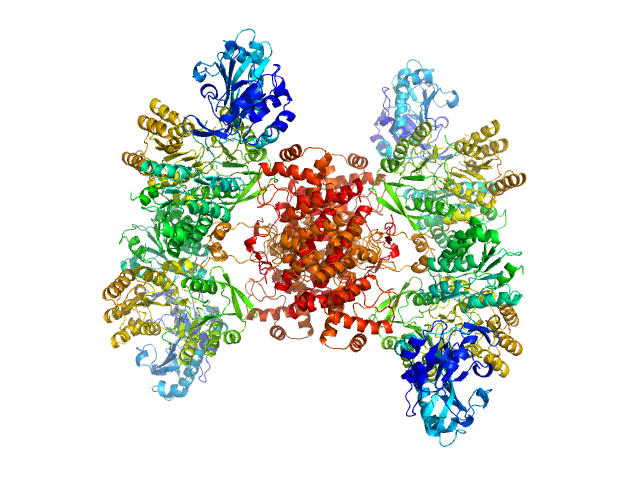
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
30.4 |
nm |
| VolumePorod |
1100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 6
|
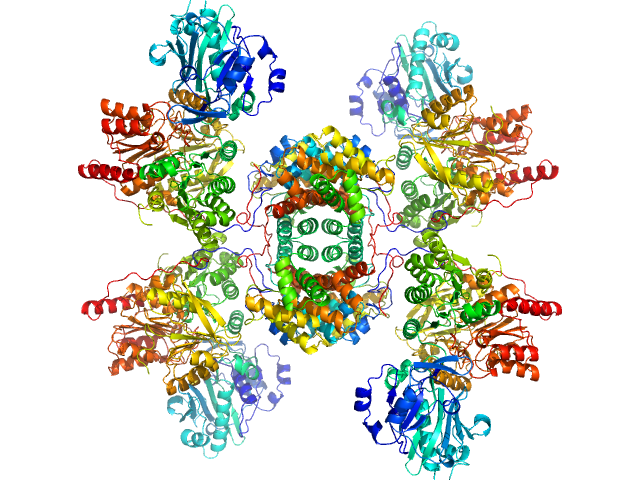
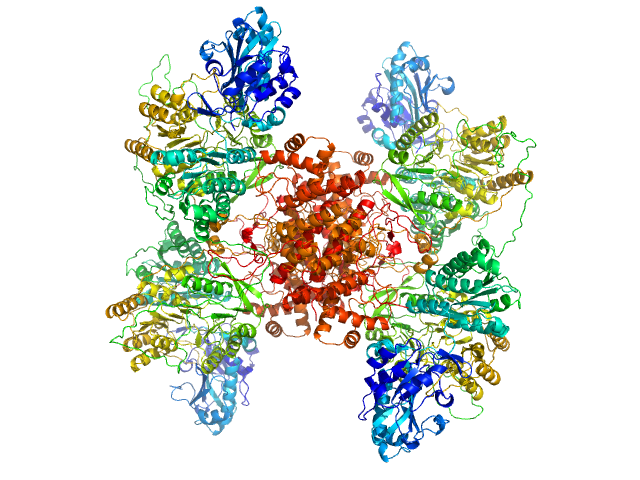
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.0 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
738 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 6
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
747 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 5
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
5.8 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
709 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.1 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
765 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.2 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
787 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
5.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
775 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyglutamine-binding protein 1 p.Lys192Serfs*7 dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
Phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2013 Feb 15
|
Frameshift PQBP-1 mutants K192Sfs*7 and R153Sfs*41 implicated in X-linked intellectual disability form stable dimers.
J Struct Biol (2019)
Rahman SK, Okazawa H, Chen YW
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyglutamine-binding protein 1 p.Arg153Serfs*41 dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
Phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2013 Feb 15
|
Frameshift PQBP-1 mutants K192Sfs*7 and R153Sfs*41 implicated in X-linked intellectual disability form stable dimers.
J Struct Biol (2019)
Rahman SK, Okazawa H, Chen YW
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Metaphase chromosomes in 17 mM magnesium chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, 17 mM magnesium chloride, 20 mM sodium chloride, 120 mM potassium chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 May 23
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|