|
|
|
|

|
| Sample: |
Bromodomain-containing protein 2 monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira N, Wilson MD, Filippakopoulos P, Gingras AC
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bromodomain testis-specific protein monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira N, Wilson MD, Filippakopoulos P, Gingras AC
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
200 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gamma-crystallin S dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2018 Feb 23
|
The structure and stability of the disulfide-linked γS-crystallin dimer provide insight into oxidation products associated with lens cataract formation.
J Mol Biol (2018)
Thorn DC, Grosas AB, Mabbitt PD, Ray NJ, Jackson CJ, Carver JA
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gamma-crystallin S monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2018 Feb 23
|
The structure and stability of the disulfide-linked γS-crystallin dimer provide insight into oxidation products associated with lens cataract formation.
J Mol Biol (2018)
Thorn DC, Grosas AB, Mabbitt PD, Ray NJ, Jackson CJ, Carver JA
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
|
|
|
|
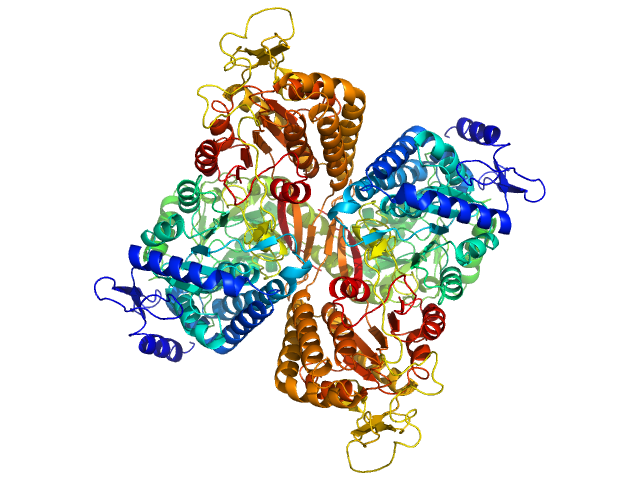
|
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
|
|
|
|
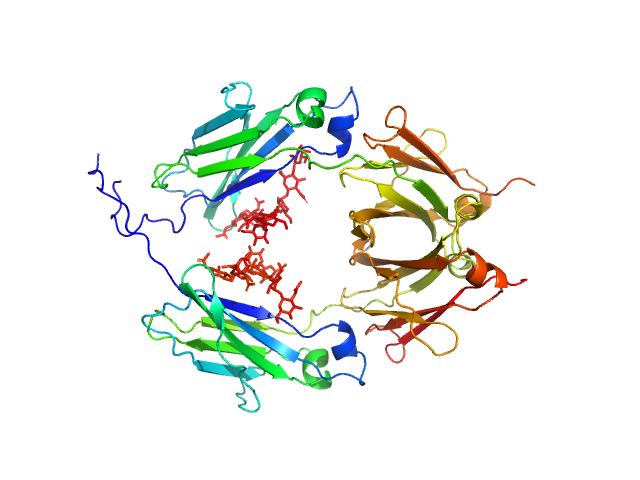
|
| Sample: |
Glycosylated human immunoglobulin G Fc region dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Citrate-Phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Mar 5
|
CH2 domain orientation of human immunoglobulin G in solution: Structural comparison of glycosylated and aglycosylated Fc regions using small-angle X-ray scattering.
MAbs (2018)
Yageta S, Imamura H, Shibuya R, Honda S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
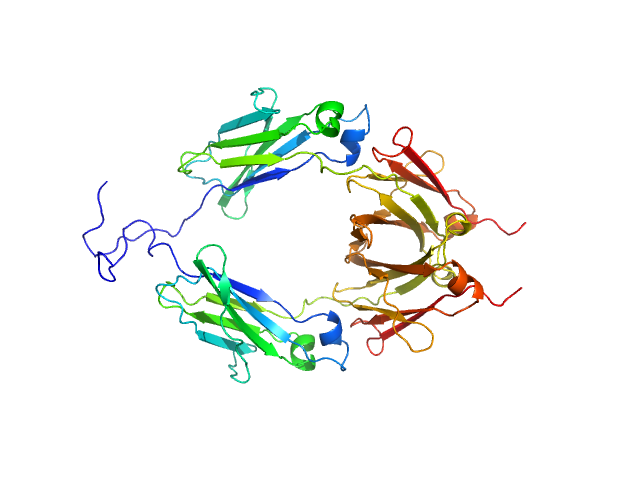
|
| Sample: |
Aglycosylated human immunoglobulin G Fc region dimer, 51 kDa Homo sapiens protein
|
| Buffer: |
20 mM Citrate-Phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Mar 5
|
CH2 domain orientation of human immunoglobulin G in solution: Structural comparison of glycosylated and aglycosylated Fc regions using small-angle X-ray scattering.
MAbs (2018)
Yageta S, Imamura H, Shibuya R, Honda S
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
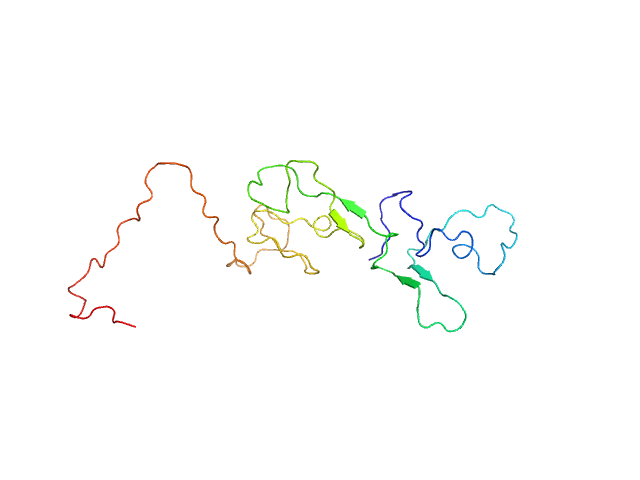
|
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 0.05 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2017 Jun 12
|
A Metastable Contact and Structural Disorder in the Estrogen Receptor Transactivation Domain.
Structure 27(2):229-240.e4 (2019)
Peng Y, Cao S, Kiselar J, Xiao X, Du Z, Hsieh A, Ko S, Chen Y, Agrawal P, Zheng W, Shi W, Jiang W, Yang L, Chance MR, Surewicz WK, Buck M, Yang S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
|
|