|
|
|
|
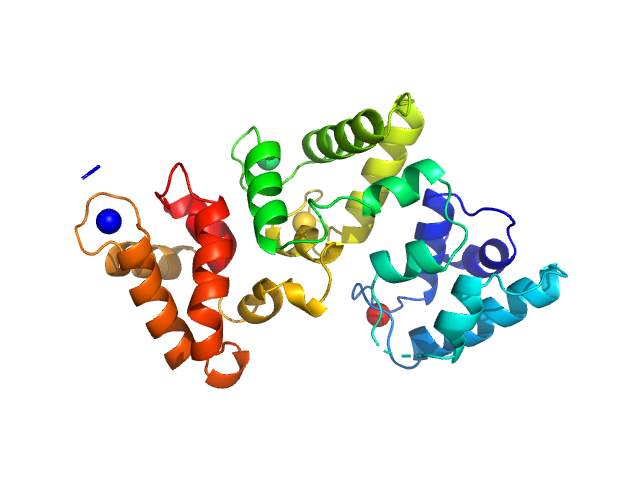
|
| Sample: |
Calbindin monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 28
|
The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol 74(Pt 10):1008-1014 (2018)
Noble JW, Almalki R, Roe SM, Wagner A, Duman R, Atack JR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Calbindin monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 28
|
The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol 74(Pt 10):1008-1014 (2018)
Noble JW, Almalki R, Roe SM, Wagner A, Duman R, Atack JR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
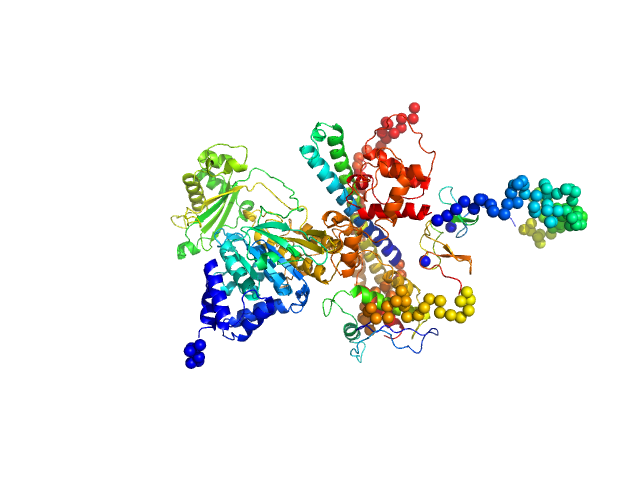
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (monomer) monomer, 118 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, pH 7.5, 5 % glycerol, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
|
|
|
|
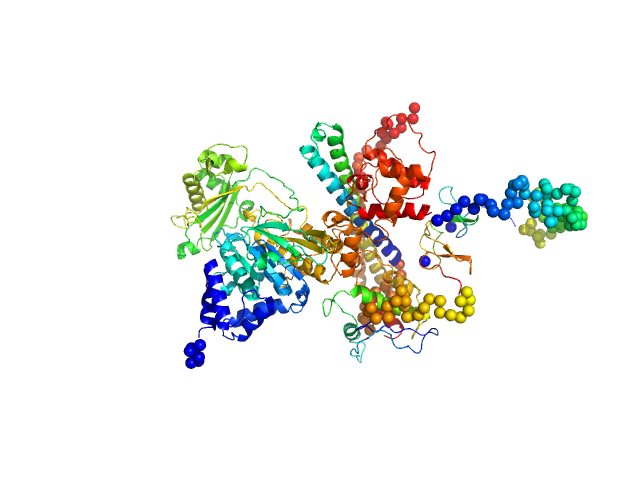
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (MoChLim) monomer, 85 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes/NaOH, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
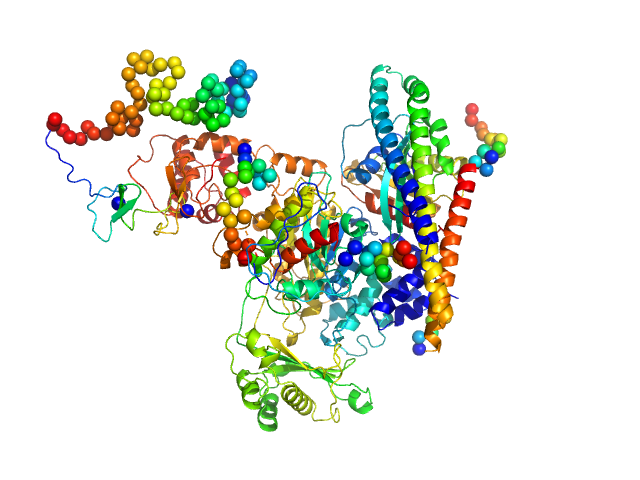
|
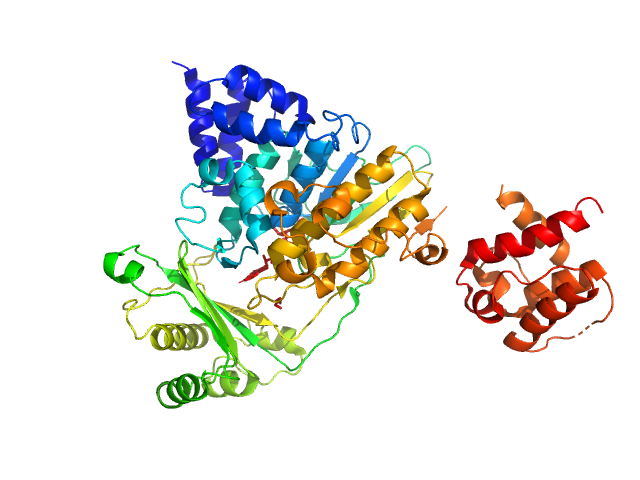
| Sample: |
[F-actin]-monooxygenase MICAL1 (monomer) monomer, 118 kDa Homo sapiens protein
Ras-related protein 8 monomer, 20 kDa protein
|
| Buffer: |
20 mM Hepes/NaOH, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
234 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (MoCh) monomer, 67 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, 5 % glycerol, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 6
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phox Homology (PX) - C2 domains of human Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris 200 mM NaCl 5% Glycerol 0.5 mM TCEP, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 20
|
Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2α.
Structure (2018)
Chen KE, Tillu VA, Chandra M, Collins BM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phox Homology (PX) - C2 domains of human Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris 200 mM NaCl 5% Glycerol 0.5 mM TCEP 4 mM InsP6, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 20
|
Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2α.
Structure (2018)
Chen KE, Tillu VA, Chandra M, Collins BM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
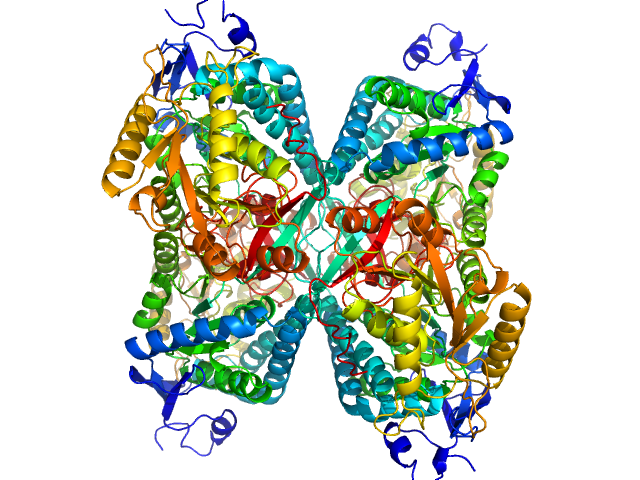
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.5 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
|
|
|
|
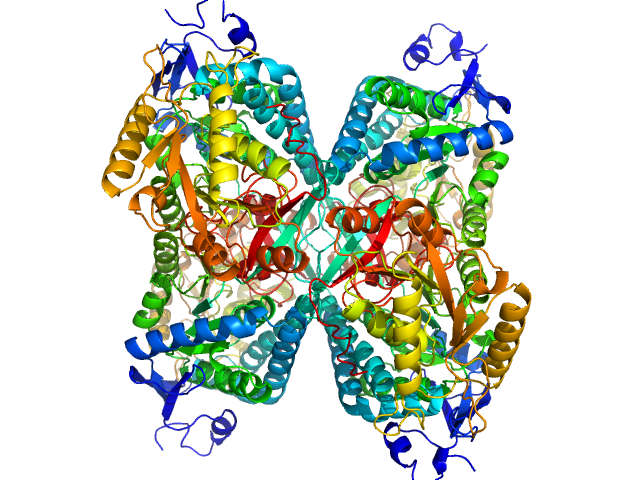
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
229 |
nm3 |
|
|