|
|
|
|
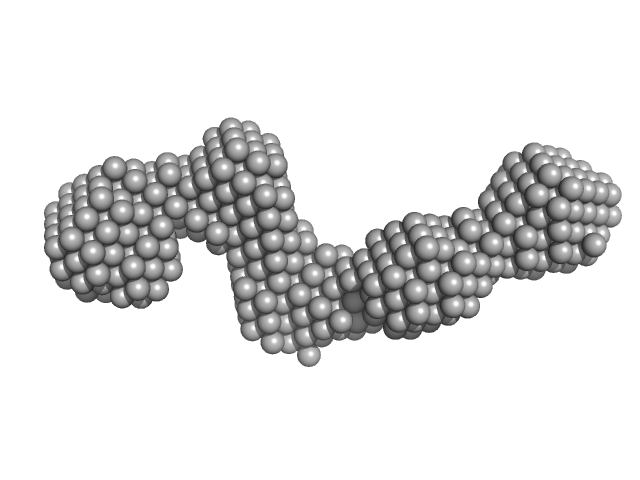
|
| Sample: |
Pre-B-cell leukemia transcription factor 1 monomer, 34 kDa Homo sapiens protein
Homeobox protein PKNOX1 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
|
|
|
|
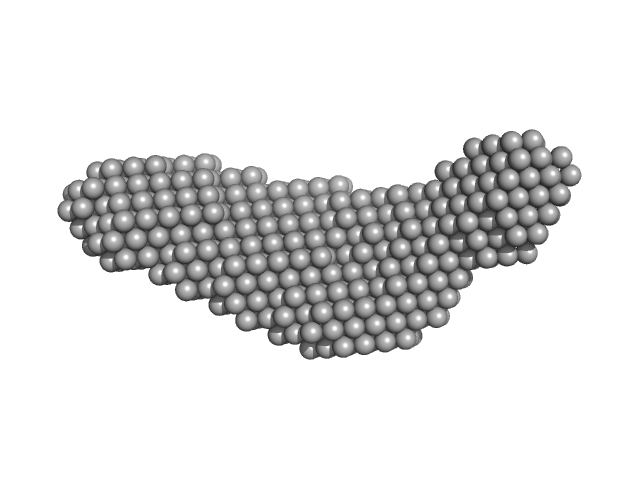
|
| Sample: |
Pre-B-cell leukemia transcription factor 1 monomer, 34 kDa Homo sapiens protein
Homeobox protein PKNOX1 monomer, 38 kDa Homo sapiens protein
DNA 22mer monomer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
|
|
|
|
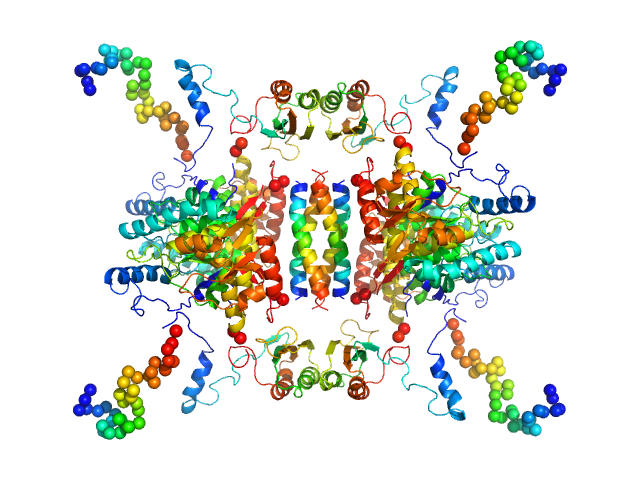
|
| Sample: |
Tyrosine hydroxylase, isoform 1 tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
Stable preparations of tyrosine hydroxylase provide the solution structure of the full-length enzyme.
Sci Rep 6:30390 (2016)
Bezem MT, Baumann A, Skjærven L, Meyer R, Kursula P, Martinez A, Flydal MI
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
520 |
nm3 |
|
|
|
|
|
|
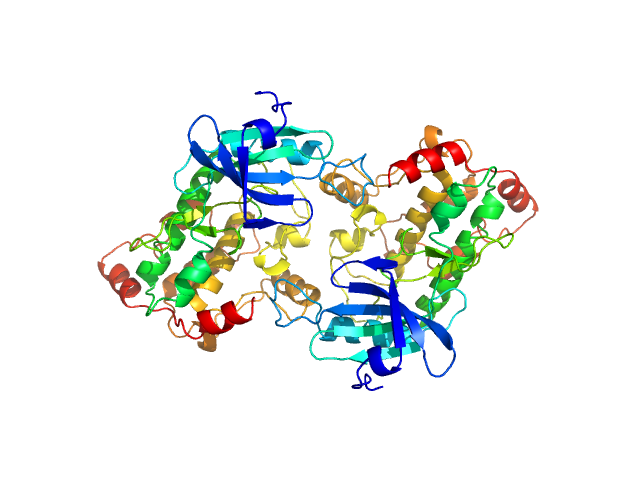
|
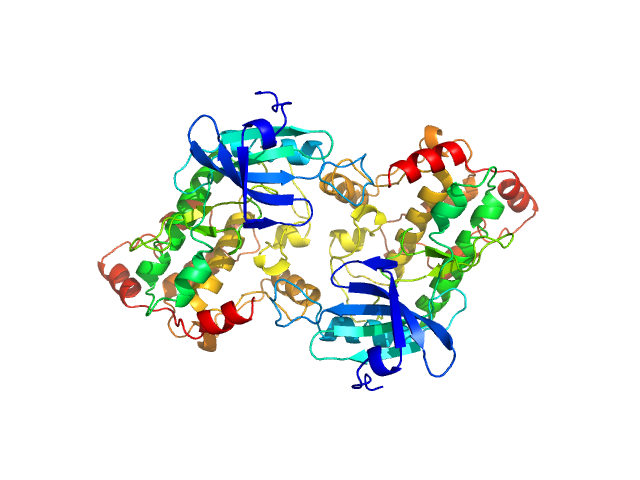
| Sample: |
Death associated protein kinase wild-type, 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
Simon B, Huart AS, Temmerman K, Vahokoski J, Mertens HD, Komadina D, Hoffmann JE, Yumerefendi H, Svergun DI, Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
|
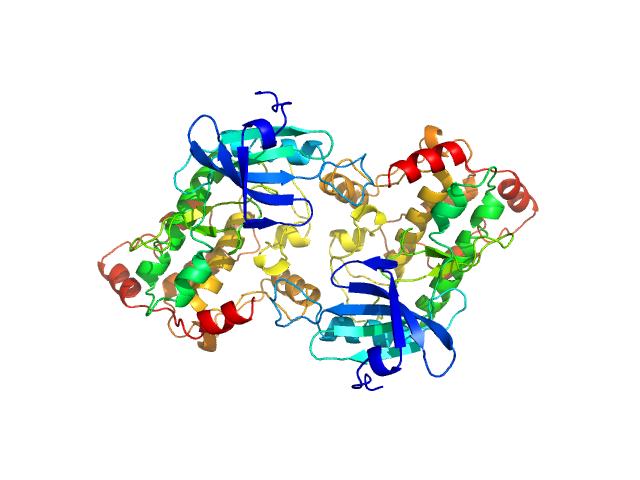
| Sample: |
Death associated protein kinase (D220K mutant), 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
Simon B, Huart AS, Temmerman K, Vahokoski J, Mertens HD, Komadina D, Hoffmann JE, Yumerefendi H, Svergun DI, Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Death associated protein kinase (Basic Loop mutant), 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
Simon B, Huart AS, Temmerman K, Vahokoski J, Mertens HD, Komadina D, Hoffmann JE, Yumerefendi H, Svergun DI, Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CDNA FLJ45612 fis, clone BRTHA3025073, highly similar to Actin cross-linking family protein 7 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 10
|
In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun 7:11692 (2016)
Yue J, Zhang Y, Liang WG, Gou X, Lee P, Liu H, Lyu W, Tang WJ, Chen SY, Yang F, Liang H, Wu X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CDNA FLJ45612 fis, clone BRTHA3025073, highly similar to Actin cross-linking family protein 7 Y259D mutant monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 7
|
In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun 7:11692 (2016)
Yue J, Zhang Y, Liang WG, Gou X, Lee P, Liu H, Lyu W, Tang WJ, Chen SY, Yang F, Liang H, Wu X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase RNF8 dimer, 35 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
4.5 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
119 |
nm3 |
|
|
|
|
|
|
|
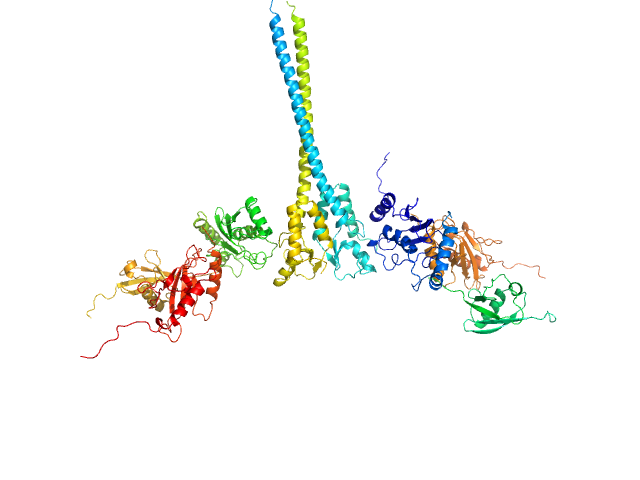
| Sample: |
E3 ubiquitin-protein ligase RNF8 dimer, 35 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 variant 2 dimer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
5.4 |
nm |
| Dmax |
23.8 |
nm |
| VolumePorod |
214 |
nm3 |
|
|