|
|
|
|
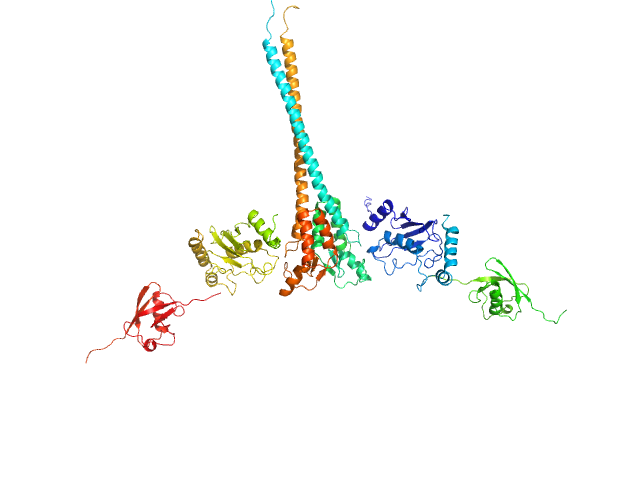
|
| Sample: |
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
E3 ubiquitin-protein ligase RNF8 mutant (L451D) dimer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
4.3 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
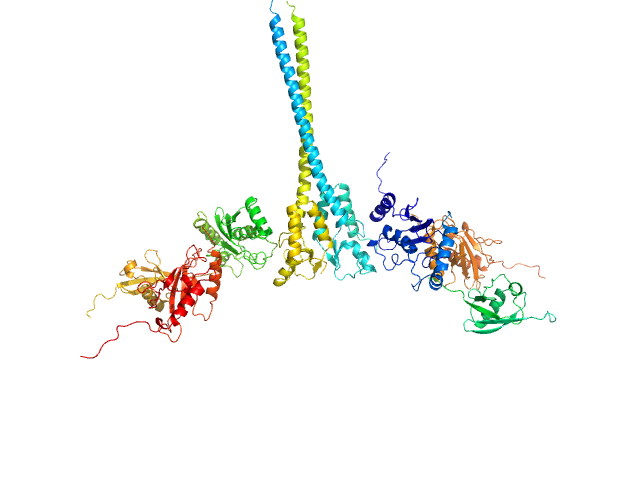
|
| Sample: |
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 variant 2 dimer, 34 kDa Homo sapiens protein
E3 ubiquitin-protein ligase RNF8 mutant (L451D) dimer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
5.2 |
nm |
| Dmax |
23.8 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Linear di-ubiquitin monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human linear tri-ubiquitin monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150mM NaCl 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human linear tetra-ubiquitin monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150mM NaCl 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
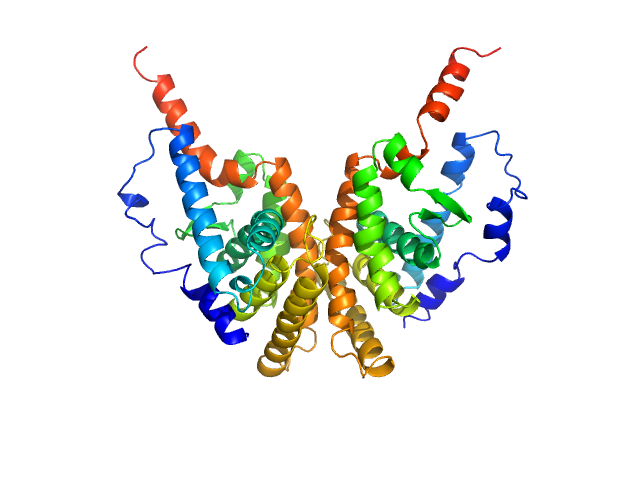
|
| Sample: |
Retinoic acid receptor RXR-alpha dimer, 51 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 100 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
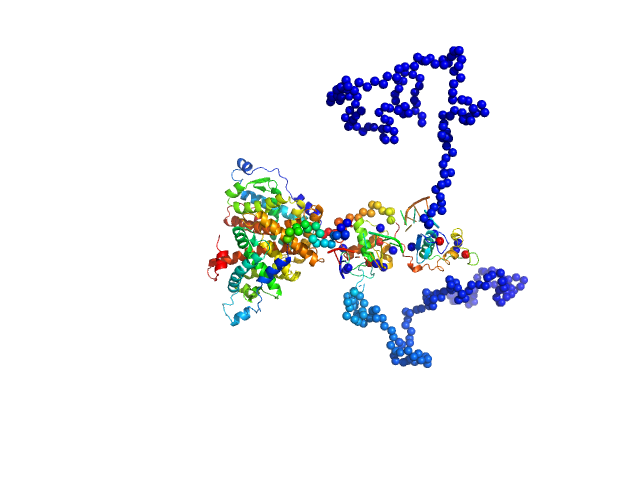
|
| Sample: |
Retinoic acid receptor RXR-alpha dimer, 102 kDa Homo sapiens protein
Ramp2 DNA monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 May 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|
|
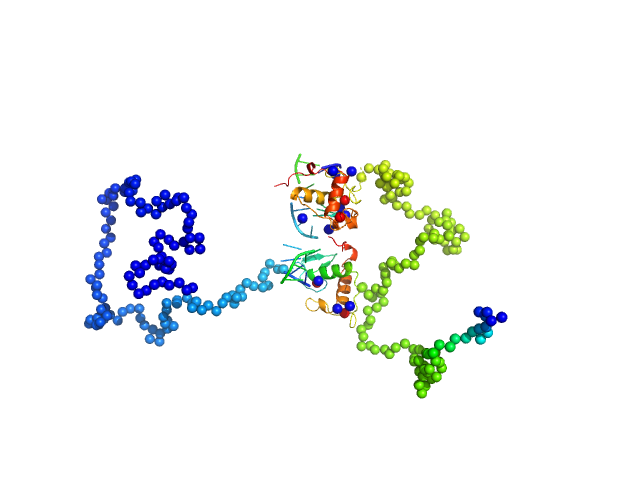
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Jul 17
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
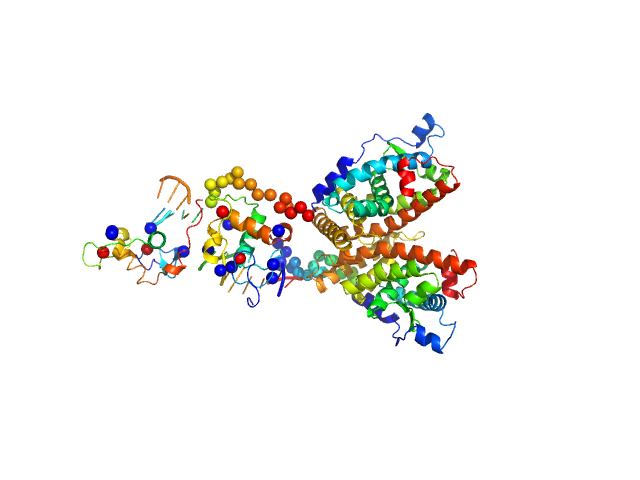
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 74 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.2 |
nm |
|
|
|
|
|
|
|
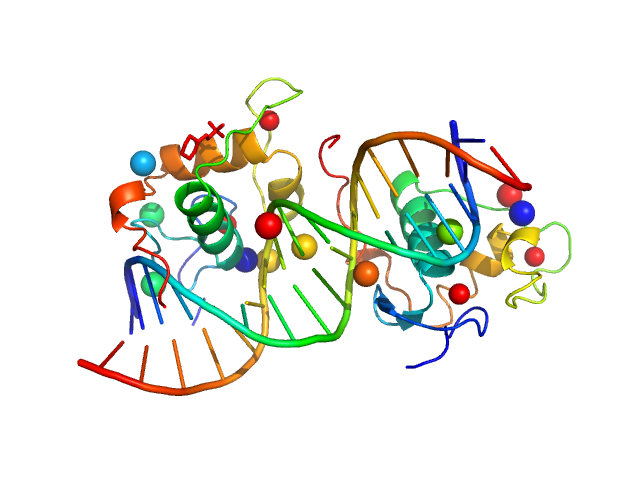
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.8 |
nm |
|
|