|
|
|
|

|
| Sample: |
Contactin-associated protein-like 2 extracellular domains (1-1261) monomer, 140 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2010 Oct 4
|
Structural Characterization of the Extracellular Domain of CASPR2 and Insights into Its Association with the Novel Ligand Contactin1.
J Biol Chem 291(11):5788-802 (2016)
Rubio-Marrero EN, Vincelli G, Jeffries CM, Shaikh TR, Pakos IS, Ranaivoson FM, von Daake S, Demeler B, De Jaco A, Perkins G, Ellisman MH, Trewhella J, Comoletti D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
282 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ankyrin repeat domains from Tankyrase 2 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human Arpin monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
2.6 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Zebrafish arpin/human tankyrase 2 ankyrin repeat domain complex monomer, 43 kDa Danio rerio / … protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cardiac myosin binding protein-C: domains C5-C6-C7 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 250 mM NaCl, 2 mM TCEP, 0.02% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 18
|
Clinically Linked Mutations in the Central Domains of Cardiac Myosin-Binding Protein C with Distinct Phenotypes Show Differential Structural Effects.
Structure 24(1):105-115 (2016)
Nadvi NA, Michie KA, Kwan AH, Guss JM, Trewhella J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human Calumenin monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
25 mM Na-HEPES, 25 mM NaCl, 2.5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Feb 12
|
Ca-Dependent Folding of Human Calumenin.
PLoS One 11(3):e0151547 (2016)
Mazzorana M, Hussain R, Sorensen T
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
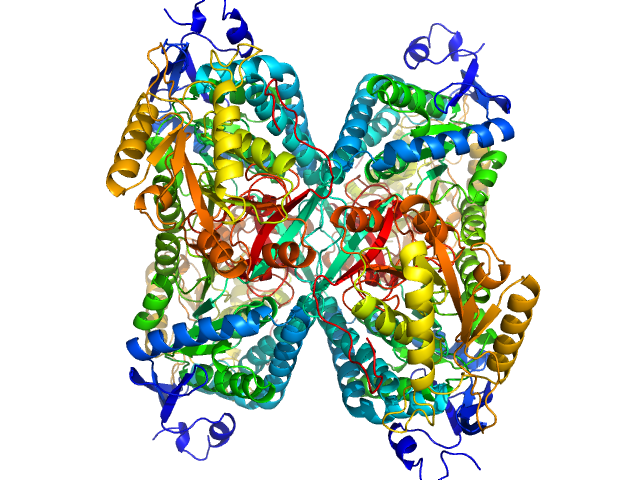
|
| Sample: |
Aldehyde dehydrogenase 7A1 (Alpha-aminoadipic semialdehyde dehydrogenase) tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 5% glycerol, 0.5 mM tris(3-hydroxypropyl)phosphine, 50 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Mar 9
|
Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry 54(35):5513-22 (2015)
Luo M, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
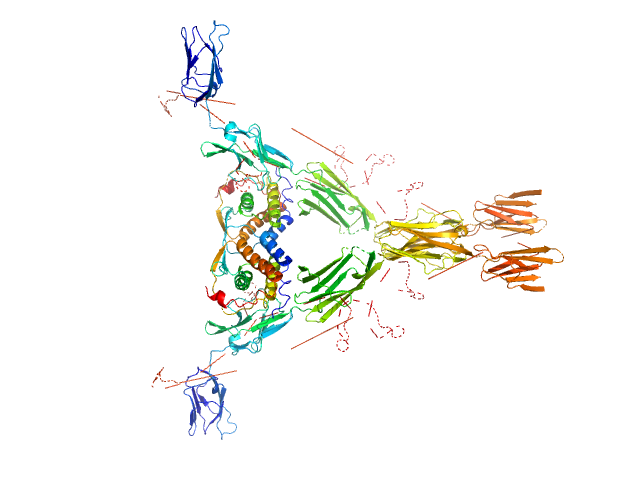
|
| Sample: |
Macrophage colony-stimulating factor 1 dimer, 35 kDa Homo sapiens protein
Macrophage colony-stimulating factor 1 receptor dimer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaH2PO4, 100 m, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 13
|
Structure and Assembly Mechanism of the Signaling Complex Mediated by Human CSF-1.
Structure 23(9):1621-1631 (2015)
Felix J, De Munck S, Verstraete K, Meuris L, Callewaert N, Elegheert J, Savvides SN
|
| RgGuinier |
5.7 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
299 |
nm3 |
|
|
|
|
|
|
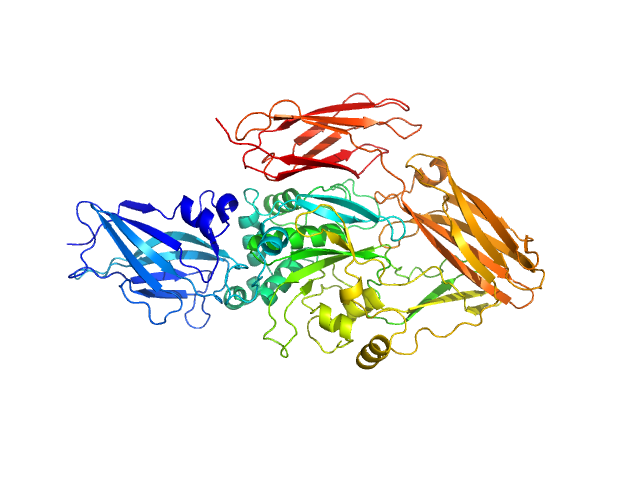
|
| Sample: |
Transglutaminase 2 monomer, 79 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150mM NaCl 1mM EDTA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jan 17
|
Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J Biol Chem 290(35):21365-75 (2015)
Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Anti-TG2 antibody (679 14 E06) monomer, 48 kDa protein
Transglutaminase 2 monomer, 79 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150mM NaCl 1mM EDTA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jan 17
|
Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J Biol Chem 290(35):21365-75 (2015)
Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
168 |
nm3 |
|
|