|
|
|
|
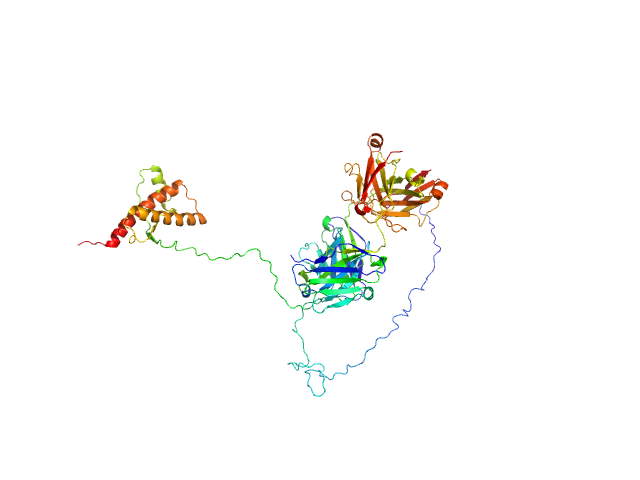
|
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
P-Clone Fab, Chimera monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
sodium acetate buffer (20 mM sodium acetate, pH 5.1; 150 mM NaCl), pH: 5.1 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2013 Dec 5
|
Prion Protein-Antibody Complexes Characterized by Chromatography-Coupled Small-Angle X-Ray Scattering.
Biophys J 109(4):793-805 (2015)
Carter L, Kim SJ, Schneidman-Duhovny D, Stöhr J, Poncet-Montange G, Weiss TM, Tsuruta H, Prusiner SB, Sali A
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hyaluronate binding domain of CD44 antigen monomer, 18 kDa Homo sapiens protein
Single-chain Variable Fragment of Antibody MEM-85 monomer, 29 kDa Mus musculus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Molecular mechanism for the action of the anti-CD44 monoclonal antibody MEM-85.
J Struct Biol 191(2):214-23 (2015)
Škerlová J, Král V, Kachala M, Fábry M, Bumba L, Svergun DI, Tošner Z, Veverka V, Řezáčová P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
100mM NaCl, 10mM sodium acetate, pH: 6 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Mar 24
|
Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife 4 (2015)
Liu Z, Gong Z, Jiang WX, Yang J, Zhu WK, Guo DC, Zhang WP, Liu ML, Tang C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
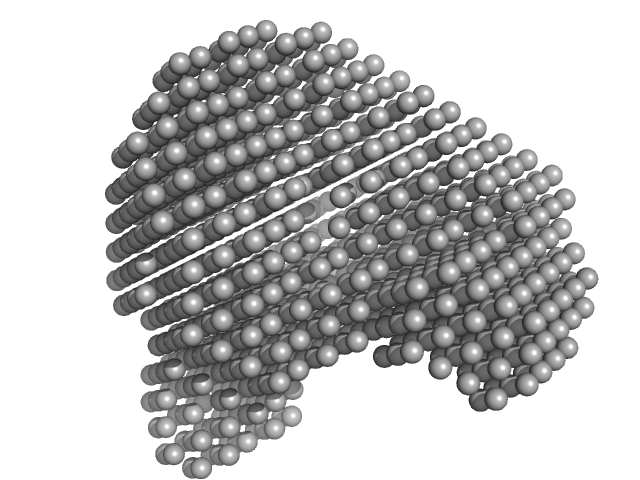
|
| Sample: |
Human serum albumin monomer monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 22
|
Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra.
Nat Methods 12(5):419-22 (2015)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Integrin beta-4 monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 6
|
Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin α6β4.
Acta Crystallogr D Biol Crystallogr 71(Pt 4):969-85 (2015)
Alonso-García N, García-Rubio I, Manso JA, Buey RM, Urien H, Sonnenberg A, Jeschke G, de Pereda JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
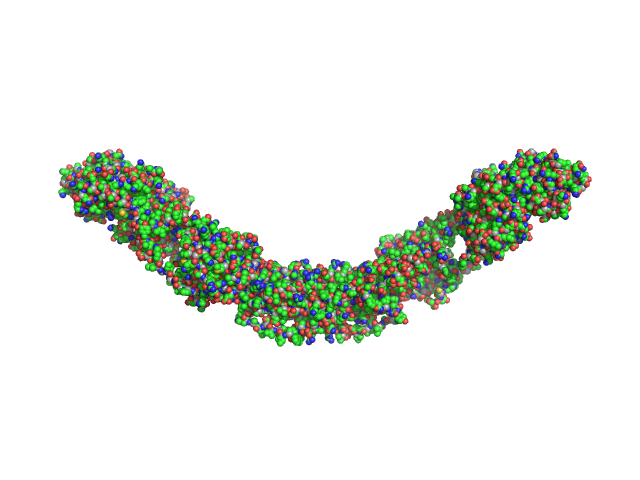
|
| Sample: |
Maltose Binding Protein fused to Protein Interacting with C kinase 1 dimer, 174 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 300 mM NaCl 1 mM maltose 1 mM EGTA 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 13
|
PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner.
Mol Biol Cell 26(7):1308-22 (2015)
Madasu Y, Yang C, Boczkowska M, Bethoney KA, Zwolak A, Rebowski G, Svitkina T, Dominguez R
|
| RgGuinier |
8.4 |
nm |
| Dmax |
27.6 |
nm |
| VolumePorod |
483 |
nm3 |
|
|
|
|
|
|
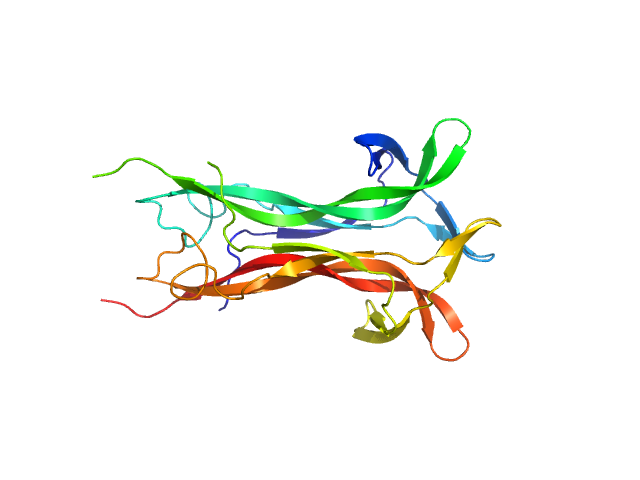
|
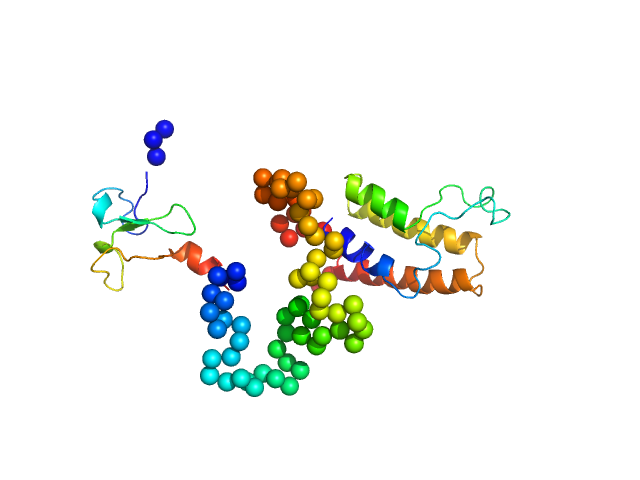
| Sample: |
Beta-nerve growth factor dimer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na-phosphate, 1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Jul 2
|
The conundrum of the high-affinity NGF binding site formation unveiled?
Biophys J 108(3):687-97 (2015)
Covaceuszach S, Konarev PV, Cassetta A, Paoletti F, Svergun DI, Lamba D, Cattaneo A
|
|
|
|
|
|
|
|
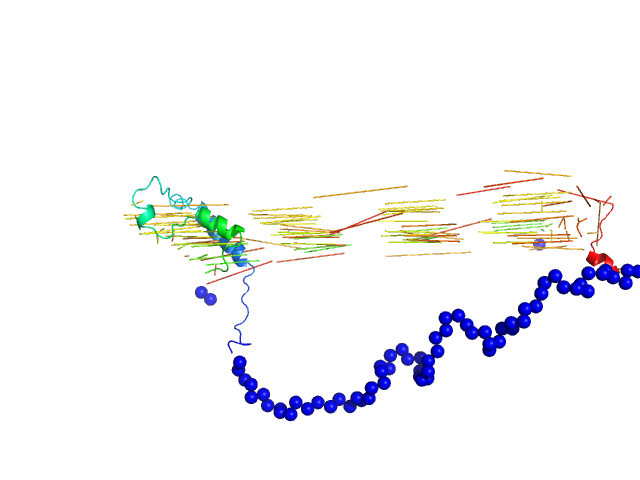
| Sample: |
Bromodomain adjacent to zinc finger domain protein 2A monomer, 27 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 500 mM NaCl 2 mM DTT 10 microM ZnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 16
|
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 23(1):80-92 (2015)
Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bromodomain adjacent to zinc finger domain protein 2B, C-terminal monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 500 mM NaCl 2 mM DTT 10 microM ZnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 24
|
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 23(1):80-92 (2015)
Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bromodomain adjacent to zinc finger domain protein 2B, C-terminal monomer, 28 kDa Homo sapiens protein
H3Kac9Kac14 monomer, 5 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 500 mM NaCl 2 mM DTT 10 microM ZnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 24
|
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 23(1):80-92 (2015)
Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|