|
|
|
|

|
| Sample: |
Deleted in Colorectal Cancer (FN5 & FN6) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 26
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin-1 monomer, 49 kDa Homo sapiens protein
Deleted in Colorectal Cancer (FN5 & FN6) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin-1 monomer, 49 kDa Homo sapiens protein
Deleted in Colorectal Cancer (FN5 & FN6) M933R mutant monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 26
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|

|
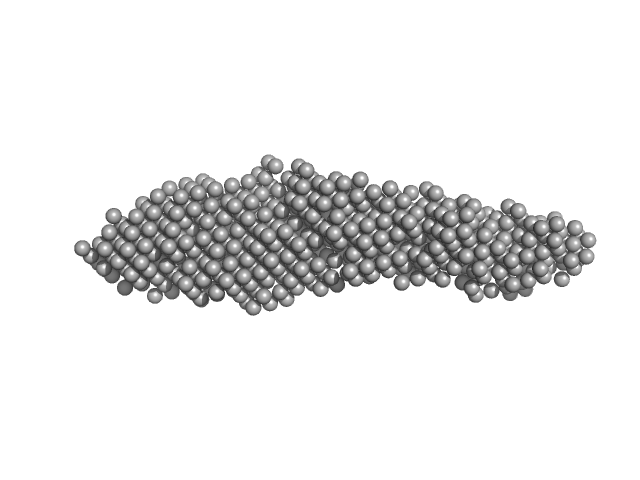
| Sample: |
Ataxin-3 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
6.9 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
425 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tubulin alpha-1A chain monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
7.0 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ataxin-3 monomer, 41 kDa Homo sapiens protein
Tubulin alpha-1A chain monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
8.4 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
900 |
nm3 |
|
|
|
|
|
|
|
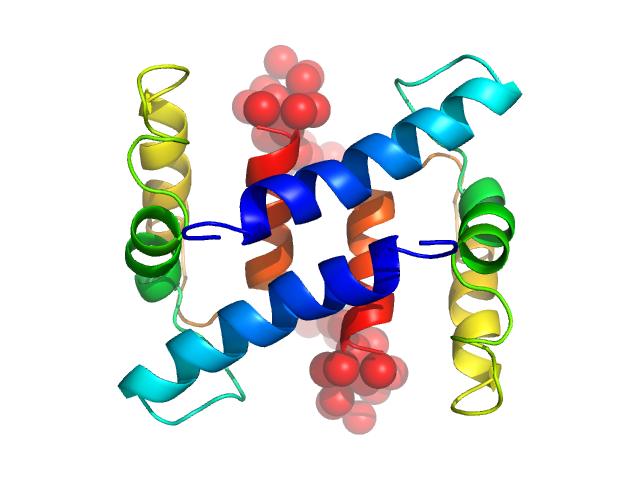
| Sample: |
Protein S100-A4 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 20 mM NaCl, 0.1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Mar 2
|
The C-Terminal Random Coil Region Tunes the Ca2+-Binding Affinity of S100A4 through Conformational Activation
PLoS ONE 9(5):e97654 (2014)
Duelli A, Kiss B, Lundholm I, Bodor A, Petoukhov M, Svergun D, Nyitray L, Katona G, Csernoch L
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.8 |
nm |
|
|