|
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆186-231) monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.6 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆362-409) monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆274-306) monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.6 |
nm |
| Dmax |
24.2 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆186-231, ∆362-409) monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.9 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform 2 of B-cell CLL/lymphoma 7 protein family member A monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.8 |
nm |
| Dmax |
28.8 |
nm |
|
|
|
|
|
|
|
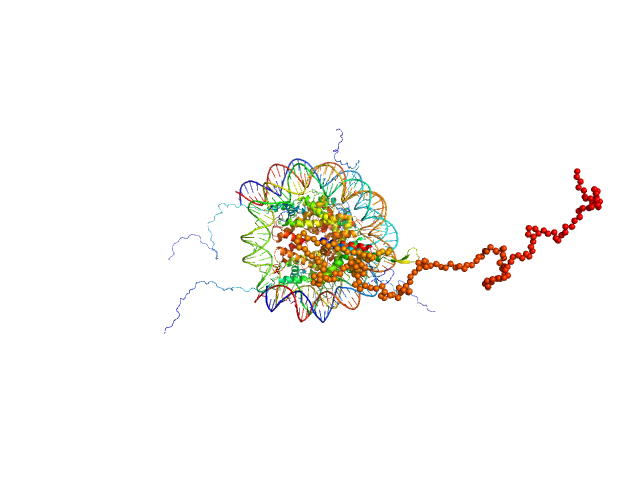
| Sample: |
Isoform 2 of B-cell CLL/lymphoma 7 protein family member A monomer, 25 kDa Homo sapiens protein
Histone H3.2 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 23 kDa Xenopus laevis protein
Histone H2A dimer, 28 kDa Xenopus laevis protein
Histone H2B 1.1 dimer, 28 kDa Xenopus laevis protein
Widom 601 DNA sequence dimer, 89 kDa Escherichia coli DNA
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.7 |
nm |
| Dmax |
24.4 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
|
|
|
|
|
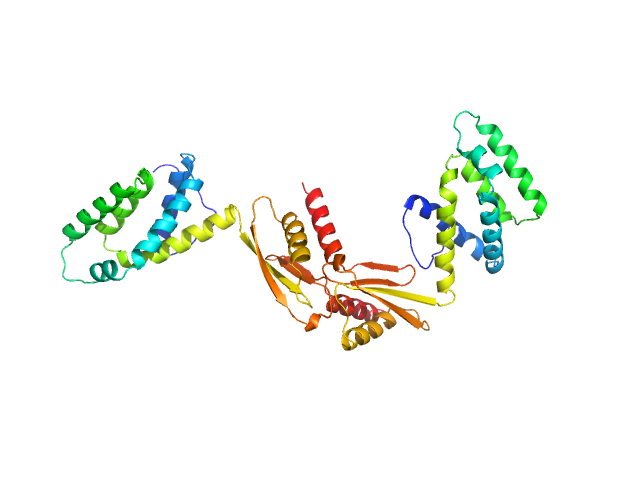
| Sample: |
ATP synthase subunit O, mitochondrial dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Mar 15
|
The OSCP Subunit of ATP Synthase is a Dimer in Solution: Strategy to Induce the Monomeric Protein as a New Tool for Drug Discovery.
J Mol Biol 437(17):169267 (2025)
Fabbian S, Gabbatore L, Morbiato L, Zotti M, Giorgio V, Battisutta R, Giachin G, Sosic A, Bellanda M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
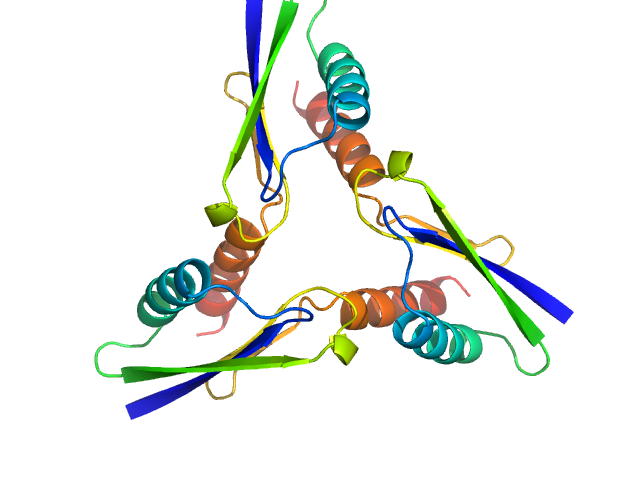
| Sample: |
ATP synthase subunit O, mitochondrial C-terminal section trimer, 25 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Mar 15
|
The OSCP Subunit of ATP Synthase is a Dimer in Solution: Strategy to Induce the Monomeric Protein as a New Tool for Drug Discovery.
J Mol Biol 437(17):169267 (2025)
Fabbian S, Gabbatore L, Morbiato L, Zotti M, Giorgio V, Battisutta R, Giachin G, Sosic A, Bellanda M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
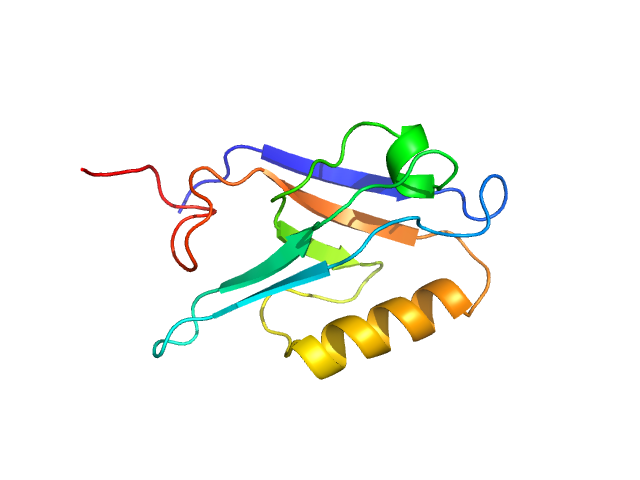
| Sample: |
Segment polarity protein dishevelled homolog DVL-3 monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate, 50 mM NaCl, 0.5 mM EDTA, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2021 Jan 27
|
A class III ligand oscillates between internal and terminal binding modes as it engages with the Dishevelled PDZ domain.
Structure (2025)
Kumar J, Micka M, Komárek J, Klumpler T, Bystrý V, Sprangers R, Bařinka C, Bryja V, Tripsianes K
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
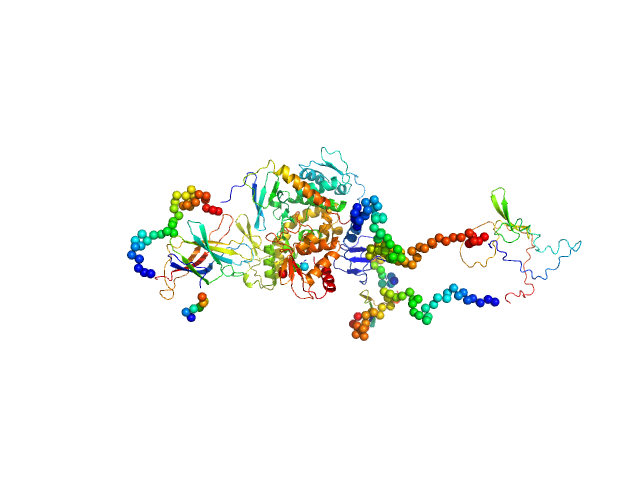
|
| Sample: |
Isoform 5 of E3 ubiquitin-protein ligase NEDD4-like monomer, 110 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 1 mM EDTA, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Aug 17
|
Structural basis of ubiquitin ligase Nedd4-2 autoinhibition and regulation by calcium and 14-3-3 proteins
Nature Communications 16(1) (2025)
Janosev M, Kosek D, Tekel A, Joshi R, Honzejkova K, Pohl P, Obsil T, Obsilova V
|
| RgGuinier |
4.9 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
206 |
nm3 |
|
|