|
|
|
|
|
| Sample: |
Isoform 5 of E3 ubiquitin-protein ligase NEDD4-like monomer, 110 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 1 mM CaCl2, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Aug 17
|
Structural basis of ubiquitin ligase Nedd4-2 autoinhibition and regulation by calcium and 14-3-3 proteins
Nature Communications 16(1) (2025)
Janosev M, Kosek D, Tekel A, Joshi R, Honzejkova K, Pohl P, Obsil T, Obsilova V
|
| RgGuinier |
4.9 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
|
|
|
|
|
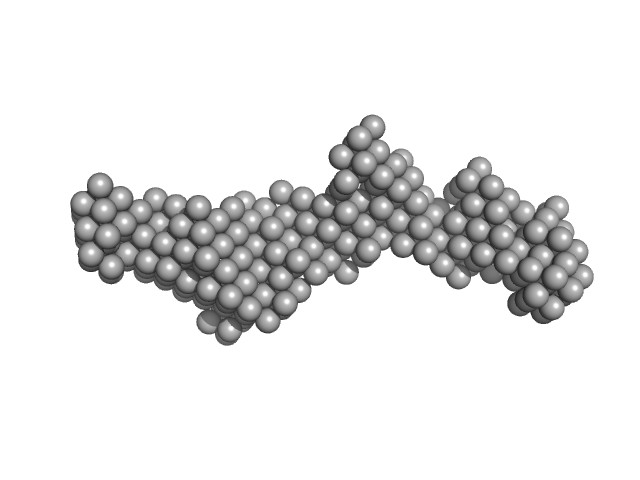
| Sample: |
Aminoacyl tRNA synthase complex-interacting multifunctional protein 1 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2022 Oct 12
|
Solution structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex
Michal Taube
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
BC120 G25C RNA monomer, 39 kDa Homo sapiens RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (L353P) dimer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 7
|
The CRISPR-Cas9 knockout DDC SH-SY5Y in vitro model for AADC deficiency provides insight into the pathogenicity of R347Q and L353P variants: a cross-sectional structural and functional analysis.
FEBS J (2025)
Carmona-Carmona CA, Bisello G, Franchini R, Lunardi G, Galavotti R, Perduca M, Ribeiro RP, Belviso BD, Giorgetti A, Caliandro R, Lievens PM, Bertoldi M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|

|
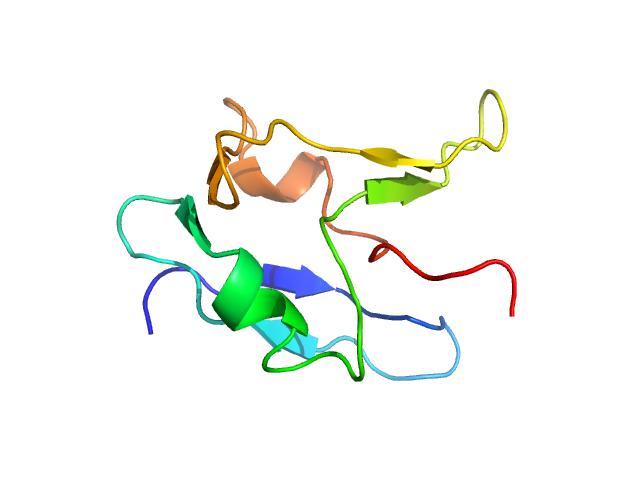
| Sample: |
Aromatic-L-amino-acid decarboxylase (R347Q) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 µM pyridoxal 5'-phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Jul 7
|
The CRISPR-Cas9 knockout DDC SH-SY5Y in vitro model for AADC deficiency provides insight into the pathogenicity of R347Q and L353P variants: a cross-sectional structural and functional analysis.
FEBS J (2025)
Carmona-Carmona CA, Bisello G, Franchini R, Lunardi G, Galavotti R, Perduca M, Ribeiro RP, Belviso BD, Giorgetti A, Caliandro R, Lievens PM, Bertoldi M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
|
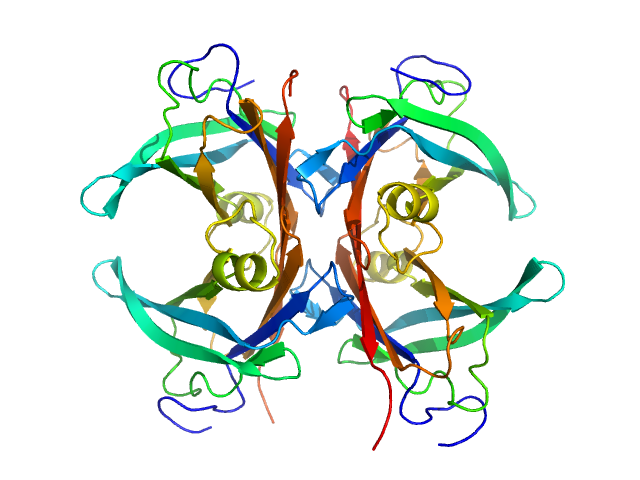
| Sample: |
TAR DNA-binding protein 43 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 3% glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BL38B1, SPring-8 on 2024 May 29
|
Semi-synthesis of TDP-43 reveals the effects of phosphorylation in N-terminal domain on self-association
Communications Chemistry 8(1) (2025)
Sasaki D, Tenda M, Sohma Y
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
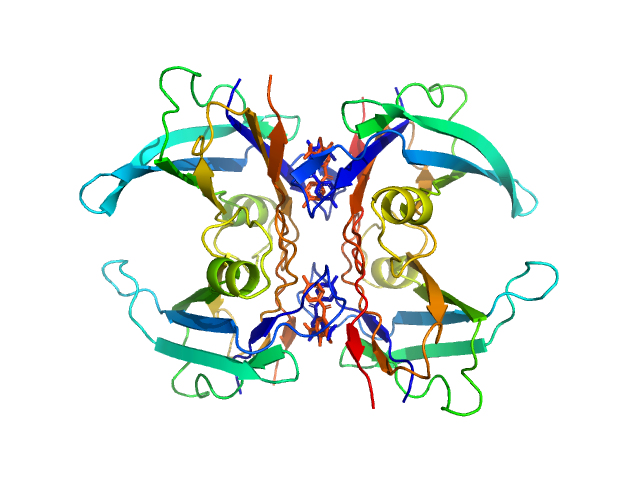
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Sep 4
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR
Orion Shih
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
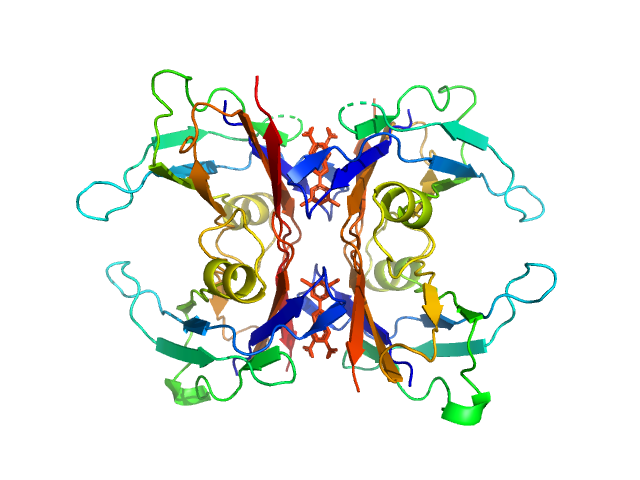
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
Tolcapone dimer, 1 kDa
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 19
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR
Orion Shih
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
Tafamidis dimer, 1 kDa
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 19
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR
Orion Shih
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
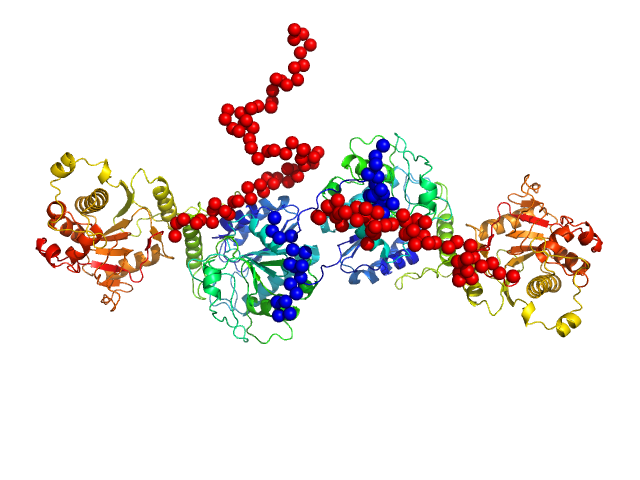
| Sample: |
Procollagen galactosyltransferase 1 dimer, 137 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 0.1 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 5
|
Molecular structure and enzymatic mechanism of the human collagen hydroxylysine galactosyltransferase GLT25D1/COLGALT1
Nature Communications 16(1) (2025)
De Marco M, Rai S, Scietti L, Mattoteia D, Liberi S, Moroni E, Pinnola A, Vetrano A, Iacobucci C, Santambrogio C, Colombo G, Forneris F
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
184 |
nm3 |
|
|