|
|
|
|
|
| Sample: |
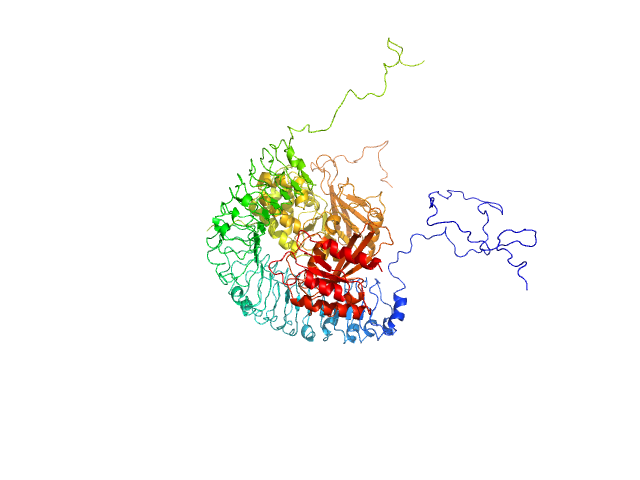
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit monomer, 37 kDa Homo sapiens protein
Ras-related protein M-Ras monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
215 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyubiquitin-B monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02% NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 8
|
Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains.
EMBO Rep :e55056 (2022)
Dao TP, Yang Y, Presti MF, Cosgrove MS, Hopkins JB, Ma W, Loh SN, Castañeda CA
|
| RgGuinier |
2.6 |
nm |
| Dmax |
95.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyubiquitin-B monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02% NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 8
|
Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains.
EMBO Rep :e55056 (2022)
Dao TP, Yang Y, Presti MF, Cosgrove MS, Hopkins JB, Ma W, Loh SN, Castañeda CA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyubiquitin-B monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02% NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 8
|
Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains.
EMBO Rep :e55056 (2022)
Dao TP, Yang Y, Presti MF, Cosgrove MS, Hopkins JB, Ma W, Loh SN, Castañeda CA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyubiquitin-B tetramer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02% NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 8
|
Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains.
EMBO Rep :e55056 (2022)
Dao TP, Yang Y, Presti MF, Cosgrove MS, Hopkins JB, Ma W, Loh SN, Castañeda CA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diacylglycerol kinase alpha monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCl, 200 mM NaCl, 5 mM DTT, 5% v/v glycerol, 3 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Dec 8
|
Ca 2+
‐induced structural changes and intramolecular interactions in N‐terminal region of diacylglycerol kinase alpha
Protein Science 31(7) (2022)
Takahashi D, Yonezawa K, Okizaki Y, Caaveiro J, Ueda T, Shimada A, Sakane F, Shimizu N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diacylglycerol kinase alpha monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20mM TrisHCl, 200mM NaCl, 5mM DTT, 5% v/v Glycerol, 3mM EGTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Dec 8
|
Ca 2+
‐induced structural changes and intramolecular interactions in N‐terminal region of diacylglycerol kinase alpha
Protein Science 31(7) (2022)
Takahashi D, Yonezawa K, Okizaki Y, Caaveiro J, Ueda T, Shimada A, Sakane F, Shimizu N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
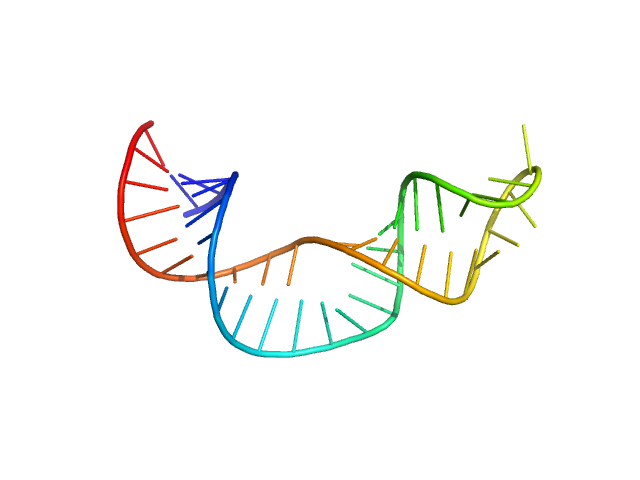
Non-pre-microRNA stem loop 2 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Solution structure of NPSL2, a regulatory element in the oncomiR-1 RNA.
J Mol Biol :167688 (2022)
Liu Y, Munsayac A, Hall I, Keane SC
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
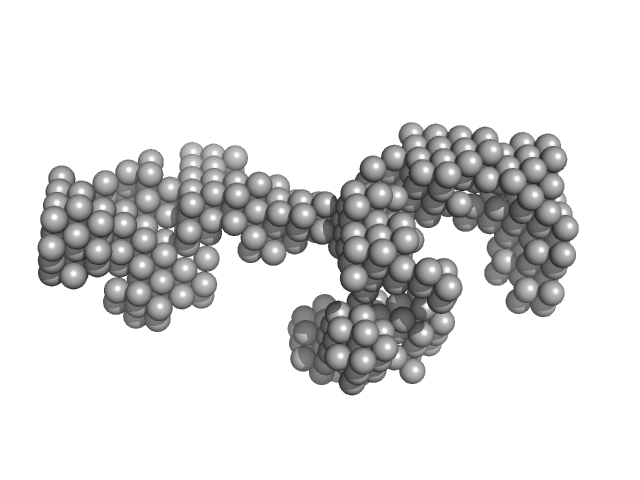
LincRNA-p21 AluSx1 Sense RNA monomer, 100 kDa Homo sapiens RNA
|
| Buffer: |
50mM HEPES,150 mM NaCl, 15 mM MgCl2, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 1
|
Biophysical characterisation of human LincRNA-p21 sense and antisense Alu inverted repeats.
Nucleic Acids Res 50(10):5881-5898 (2022)
D'Souza MH, Mrozowich T, Badmalia MD, Geeraert M, Frederickson A, Henrickson A, Demeler B, Wolfinger MT, Patel TR
|
| RgGuinier |
6.1 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LincRNA-p21 AluSx1 Antisense RNA monomer, 90 kDa Homo sapiens RNA
|
| Buffer: |
50mM HEPES,150 mM NaCl, 15 mM MgCl2, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 1
|
Biophysical characterisation of human LincRNA-p21 sense and antisense Alu inverted repeats.
Nucleic Acids Res 50(10):5881-5898 (2022)
D'Souza MH, Mrozowich T, Badmalia MD, Geeraert M, Frederickson A, Henrickson A, Demeler B, Wolfinger MT, Patel TR
|
| RgGuinier |
5.9 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
375 |
nm3 |
|
|