|
|
|
|
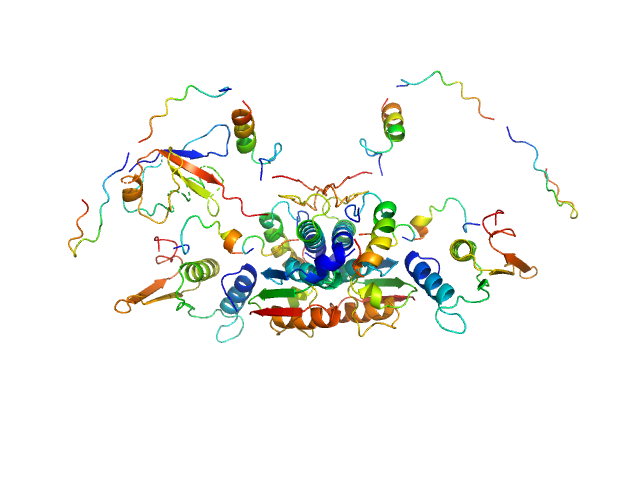
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
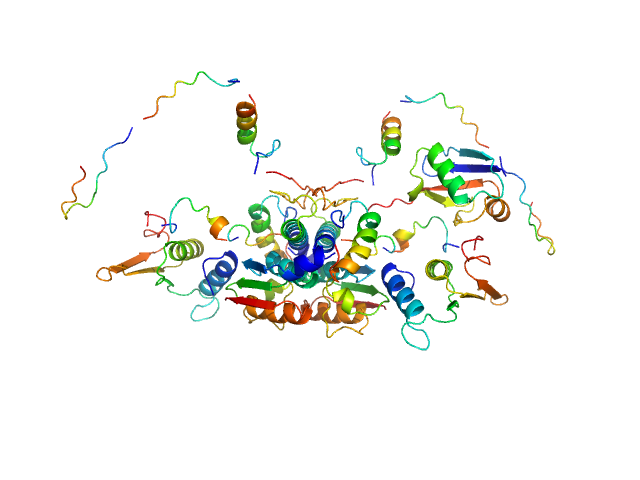
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoid-related orphan receptor-gamma monomer, 56 kDa Homo sapiens protein
Classic-RORgamma Response Element dimer, 19 kDa Homo sapiens DNA
|
| Buffer: |
25 mM HEPES, 150 mM TCEP, 2% Glycerol, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Nov 20
|
Conformational Changes of RORγ During Response Element Recognition and Coregulator Engagement
Journal of Molecular Biology :167258 (2021)
Strutzenberg T, Zhu Y, Novick S, Garcia-Ordonez R, Doebelin C, He Y, Ra Chang M, Kamenecka T, Edwards D, Griffin P
|
| RgGuinier |
5.5 |
nm |
| Dmax |
22.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoid-related orphan receptor-gamma monomer, 56 kDa Homo sapiens protein
Variant-RORgamma Response Element dimer, 18 kDa Homo sapiens DNA
|
| Buffer: |
25 mM HEPES, 150 mM TCEP, 2% Glycerol, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Nov 20
|
Conformational Changes of RORγ During Response Element Recognition and Coregulator Engagement
Journal of Molecular Biology :167258 (2021)
Strutzenberg T, Zhu Y, Novick S, Garcia-Ordonez R, Doebelin C, He Y, Ra Chang M, Kamenecka T, Edwards D, Griffin P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
22.1 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
|
|
|
|
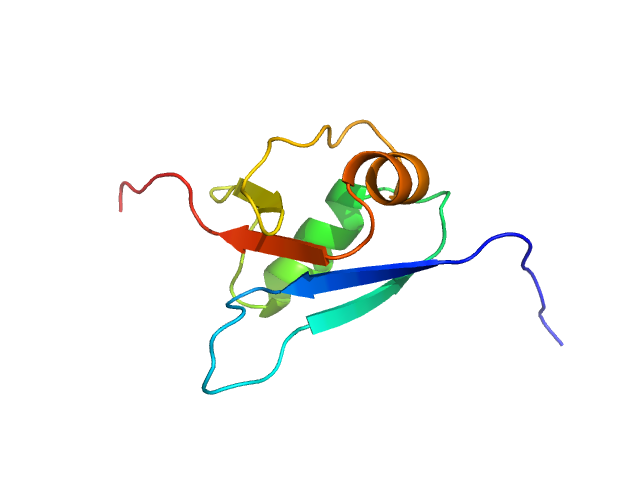
|
| Sample: |
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 13
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurogenic locus notch homolog protein 1 full ectodomain monomer, 183 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 25
|
Notch-Jagged signaling complex defined by an interaction mosaic.
Proc Natl Acad Sci U S A 118(30) (2021)
Zeronian MR, Klykov O, Portell I de Montserrat J, Konijnenberg MJ, Gaur A, Scheltema RA, Janssen BJC
|
| RgGuinier |
10.5 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
1000 |
nm3 |
|
|
|
|
|
|
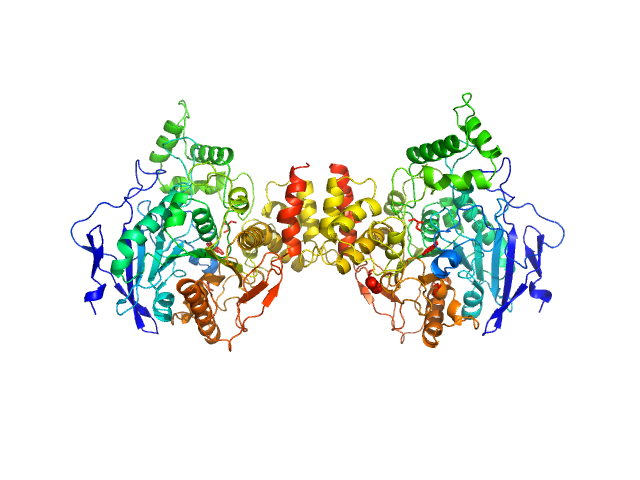
|
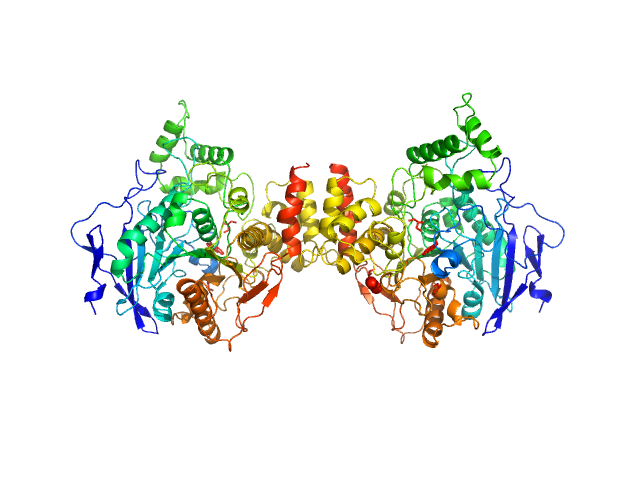
| Sample: |
Acetylcholinesterase dimer, 120 kDa Homo sapiens protein
Acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Acetylcholinesterase dimer, 120 kDa Homo sapiens protein
Acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
|
|
|
|
|
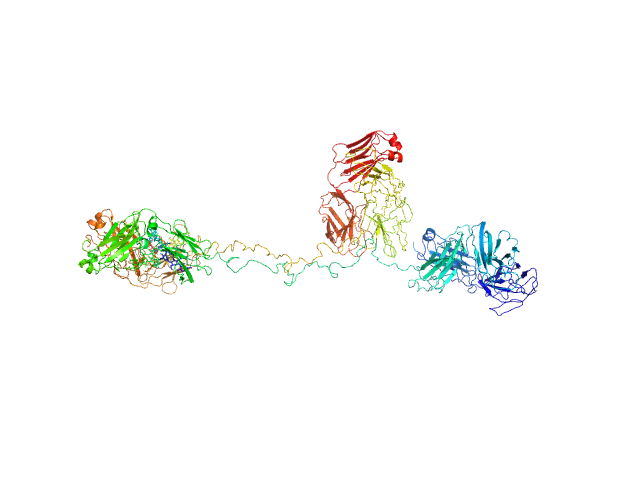
| Sample: |
Immunoglobulin G subclass 3 monomer, 155 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
Solution structures of human myeloma IgG3 antibody reveal extended Fab and Fc regions relative to the other IgG subclasses.
J Biol Chem :100995 (2021)
Spiteri VA, Goodall M, Doutch J, Rambo RP, Gor J, Perkins SJ
|
| RgGuinier |
7.0 |
nm |
| Dmax |
22.9 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
|
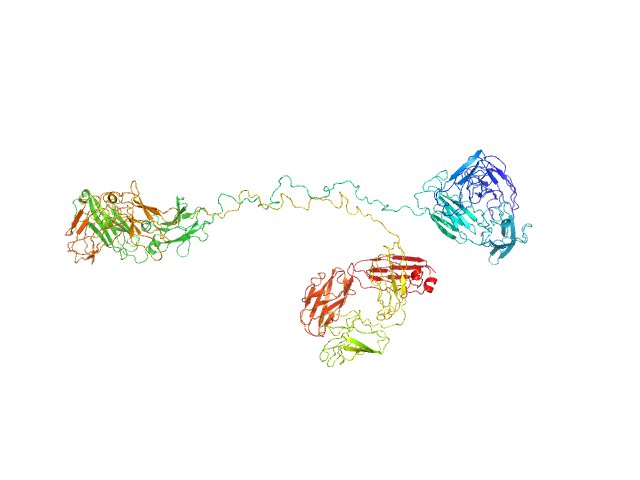
| Sample: |
Immunoglobulin G subclass 3 monomer, 155 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
Solution structures of human myeloma IgG3 antibody reveal extended Fab and Fc regions relative to the other IgG subclasses.
J Biol Chem :100995 (2021)
Spiteri VA, Goodall M, Doutch J, Rambo RP, Gor J, Perkins SJ
|
| RgGuinier |
7.1 |
nm |
| Dmax |
27.7 |
nm |
| VolumePorod |
294 |
nm3 |
|
|