|
|
|
|

|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-783 dimer, 138 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
|
|
|
|
|
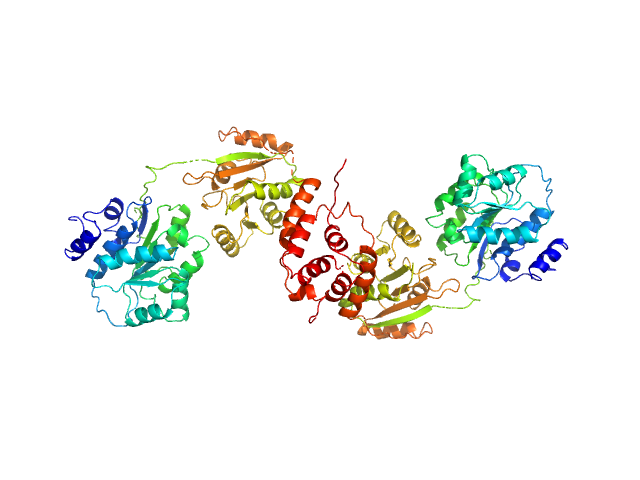
| Sample: |
Nucleolar RNA helicase 2 fragment 186-710 dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-620 dimer, 98 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolar RNA helicase 2 monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 3
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Full-length HERV-K Rec response element monomer, 141 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 10
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
7.5 |
nm |
| Dmax |
28.2 |
nm |
| VolumePorod |
454 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Truncated HERV-K Rec response element monomer, 102 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jul 17
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
322 |
nm3 |
|
|
|
|
|
|
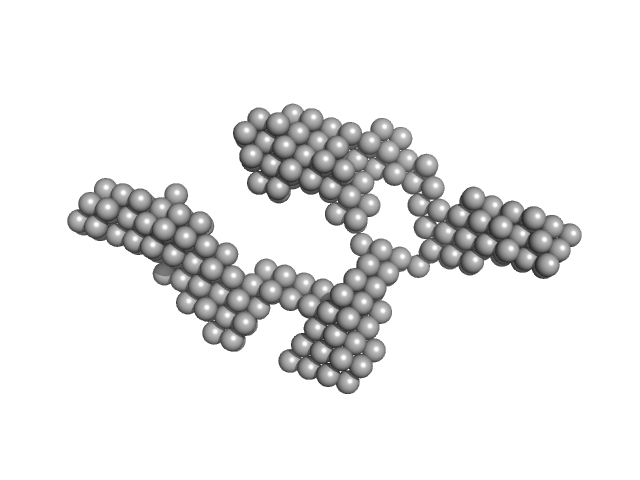
|
| Sample: |
TrΔSLIII HERV-K Rec response element monomer, 88 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Dec 7
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
5.3 |
nm |
| Dmax |
19.2 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
|
|
|
|

|
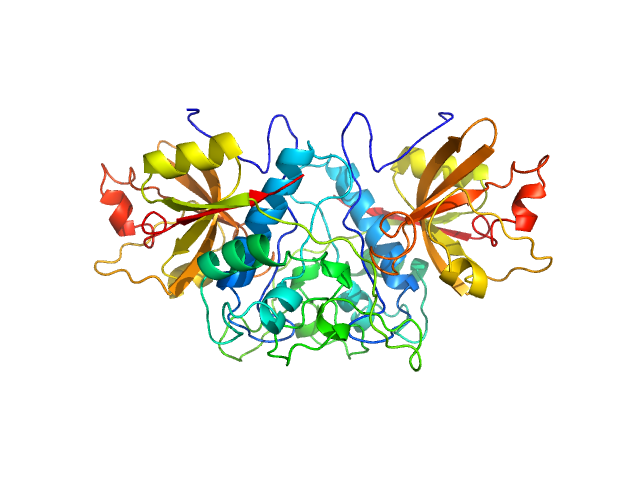
| Sample: |
TrΔSLIIab HERV-K Rec response element monomer, 86 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 21
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cathepsin Z dimer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium acetate, 1 mM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 6
|
Human cathepsin X/Z is a biologically active homodimer.
Biochim Biophys Acta Proteins Proteom :140567 (2020)
Dolenc I, Štefe I, Turk D, Taler-Verčič A, Turk B, Turk V, Stoka V
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Primary microRNA pri-miR16-1 monomer, 36 kDa Homo sapiens RNA
|
| Buffer: |
50 mM KCl, 50 mM HEPES, 5 mM DTT, 1% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Apr 12
|
Elucidating the Role of Microprocessor Protein DGCR8 in Bending RNA Structures
Biophysical Journal (2020)
Pabit S, Chen Y, Usher E, Cook E, Pollack L, Showalter S
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
74 |
nm3 |
|
|