|
|
|
|
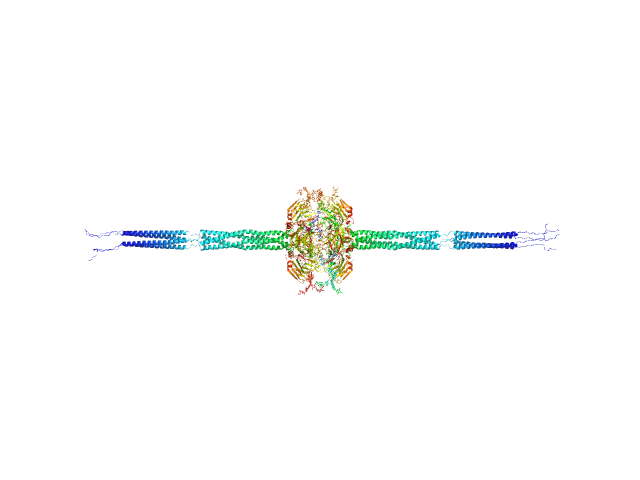
|
| Sample: |
Pentraxin-related protein PTX3 (N-terminal deletion mutant) octamer, 301 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Matrix Biology 136:52-68 (2025)
Shah A, Zhang X, Snee M, Lockhart-Cairns M, Levy C, Jowitt T, Birchenough H, Dean L, Collins R, Dodd R, Roberts A, Enghild J, Mantovani A, Fontana J, Baldock C, Inforzato A, Richter R, Day A
|
| RgGuinier |
8.4 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
647 |
nm3 |
|
|
|
|
|
|
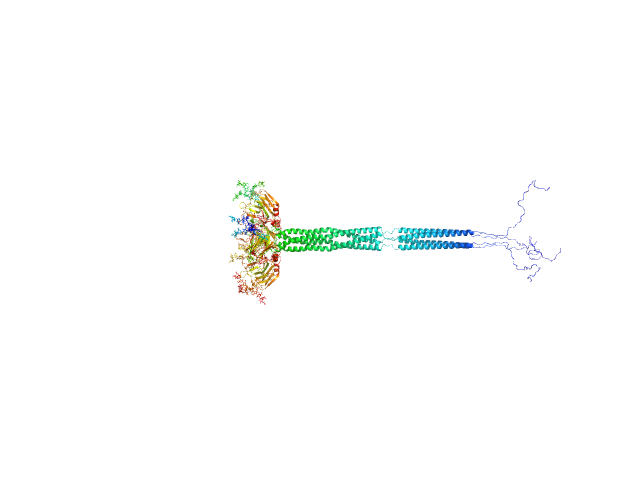
|
| Sample: |
Pentraxin-related protein PTX3 (C317S, C318S) tetramer, 160 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Matrix Biology 136:52-68 (2025)
Shah A, Zhang X, Snee M, Lockhart-Cairns M, Levy C, Jowitt T, Birchenough H, Dean L, Collins R, Dodd R, Roberts A, Enghild J, Mantovani A, Fontana J, Baldock C, Inforzato A, Richter R, Day A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDVV6_fit1_model1.png)
|
| Sample: |
Drebrin monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
17 mM NaH2PO4, 3 mM Na2HPO4, 50 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jul 7
|
Dynamic Interchange of Local Residue-Residue Interactions in the Largely Extended Single Alpha-Helix in Drebrin
Biochemical Journal (2025)
Varga S, Péterfia B, Dudola D, Farkas V, Jeffries C, Permi P, Gáspári Z
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
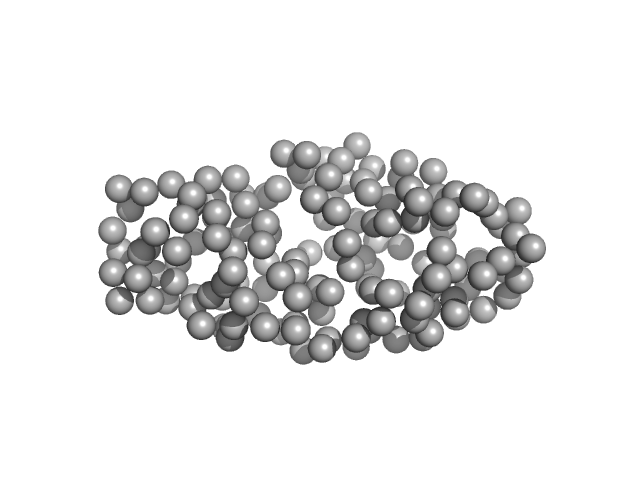
|
| Sample: |
DNA (cytosine-5)-methyltransferase 3B Pro-Trp-Trp-Pro (PWWP) domain monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2024 Apr 20
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
20464 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
271263 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Histone H3.3 dimer, 8 kDa Homo sapiens protein
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Histone H3.3 monomer, 4 kDa Homo sa protein
DNA (cytosine-5)-methyltransferase 3B Pro-Trp-Trp-Pro (PWWP) domain monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2024 Sep 17
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
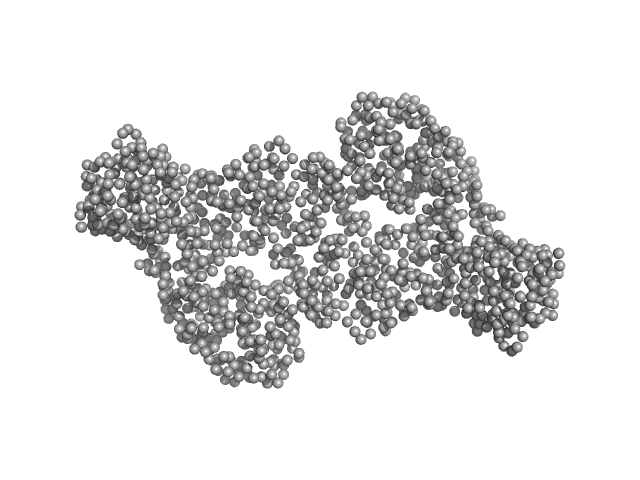
|
| Sample: |
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
DNA methyltransferase 3 beta (413-853) dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Nov 2
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
191 |
nm3 |
|
|
|
|
|
|
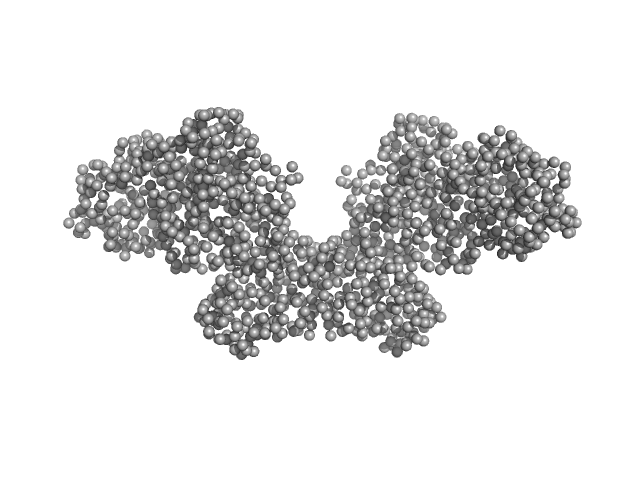
|
| Sample: |
Histone H3.3 dimer, 8 kDa Homo sapiens protein
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
DNA methyltransferase 3 beta (413-853) dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Nov 2
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
4.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Jun 2
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
117 |
nm3 |
|
|