|
|
|
|
|
| Sample: |
Myotilin Ig1Ig2 (220-452) monomer, 27 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na+-HEPES, 150 mM, NaCl, 5 % v/v glycerol, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2010 Nov 16
|
Molecular basis of F-actin regulation and sarcomere assembly via myotilin
PLOS Biology 19(4):e3001148 (2021)
Kostan J, Pavšič M, Puž V, Schwarz T, Drepper F, Molt S, Graewert M, Schreiner C, Sajko S, van der Ven P, Onipe A, Svergun D, Warscheid B, Konrat R, Fürst D, Lenarčič B, Djinović-Carugo K, Machesky L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myotilin Ig1Ig2 (250-444) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na+-HEPES, 150 mM, NaCl, 5 % v/v glycerol, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2019 Jun 27
|
Molecular basis of F-actin regulation and sarcomere assembly via myotilin
PLOS Biology 19(4):e3001148 (2021)
Kostan J, Pavšič M, Puž V, Schwarz T, Drepper F, Molt S, Graewert M, Schreiner C, Sajko S, van der Ven P, Onipe A, Svergun D, Warscheid B, Konrat R, Fürst D, Lenarčič B, Djinović-Carugo K, Machesky L
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myotilin Ig1Ig2 (250-498) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na+-HEPES, 150 mM, NaCl, 5 % v/v glycerol, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2019 Jun 27
|
Molecular basis of F-actin regulation and sarcomere assembly via myotilin
PLOS Biology 19(4):e3001148 (2021)
Kostan J, Pavšič M, Puž V, Schwarz T, Drepper F, Molt S, Graewert M, Schreiner C, Sajko S, van der Ven P, Onipe A, Svergun D, Warscheid B, Konrat R, Fürst D, Lenarčič B, Djinović-Carugo K, Machesky L
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myotilin (222-452) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 % glycerol, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jul 6
|
Molecular basis of F-actin regulation and sarcomere assembly via myotilin
PLOS Biology 19(4):e3001148 (2021)
Kostan J, Pavšič M, Puž V, Schwarz T, Drepper F, Molt S, Graewert M, Schreiner C, Sajko S, van der Ven P, Onipe A, Svergun D, Warscheid B, Konrat R, Fürst D, Lenarčič B, Djinović-Carugo K, Machesky L
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myotilin (222-452) R405K monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 % glycerol, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jul 12
|
Molecular basis of F-actin regulation and sarcomere assembly via myotilin
PLOS Biology 19(4):e3001148 (2021)
Kostan J, Pavšič M, Puž V, Schwarz T, Drepper F, Molt S, Graewert M, Schreiner C, Sajko S, van der Ven P, Onipe A, Svergun D, Warscheid B, Konrat R, Fürst D, Lenarčič B, Djinović-Carugo K, Machesky L
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
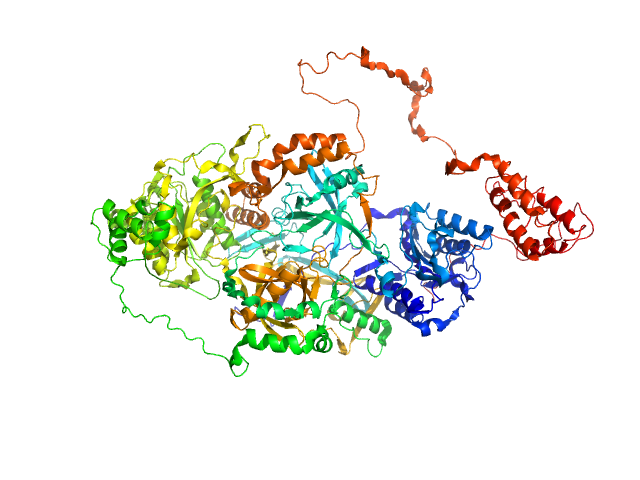
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
|
|
|
|
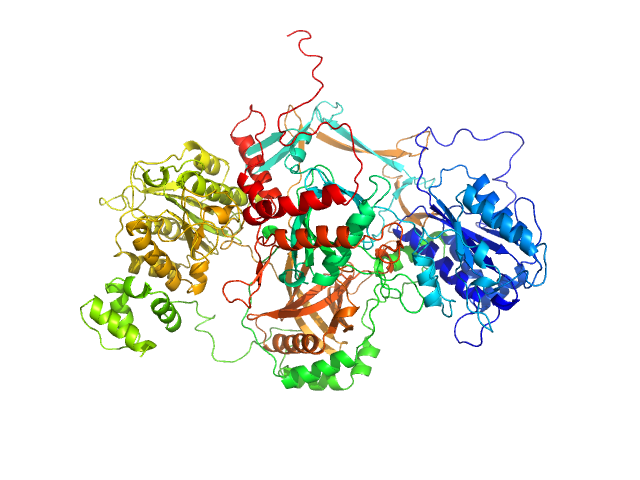
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 ΔCTR monomer, 64 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
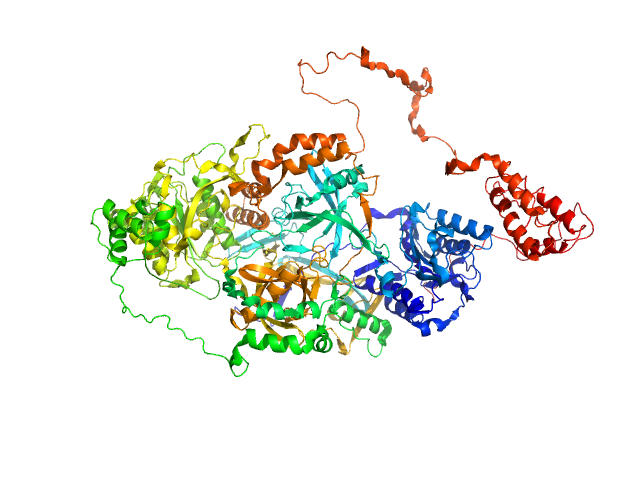
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
Y-DNA monomer, 18 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Jan 8
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
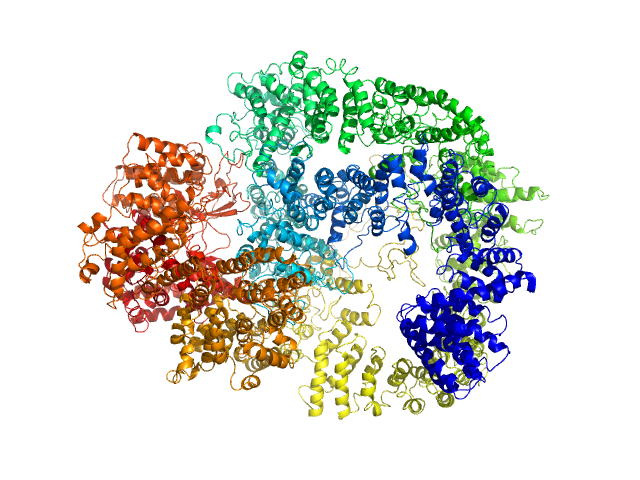
|
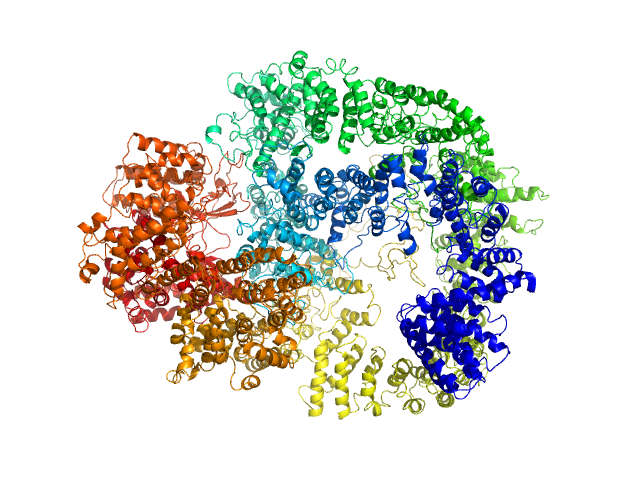
| Sample: |
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
5.6 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
1040 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM KCl, 5% (v/v) glycerol, 0.2 mM EDTA containing 0.1 mM benzamidine, 0.2 mM PMSF and 0.2 µg/ml pepstatin, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Jan 8
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
5.5 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
918 |
nm3 |
|
|