|
|
|
|
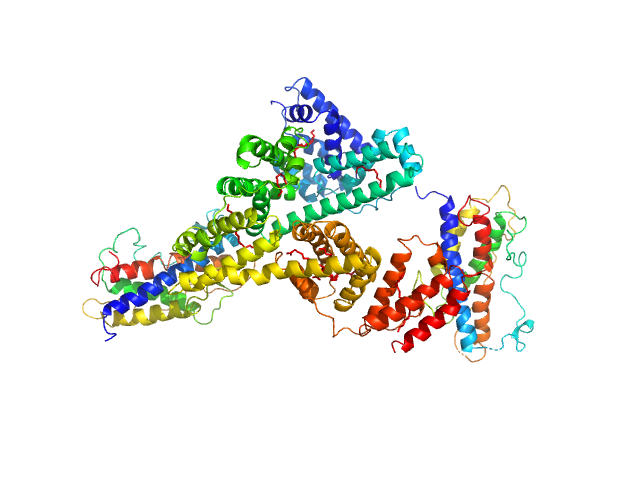
|
| Sample: |
Human serum albumin monomer, 66 kDa Homo sapiens protein
Somapacitan dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
100 mM MES, 140 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Novo Nordisk A/S on 2015 Sep 4
|
Identification of binding sites on human serum albumin for somapacitan - a long-acting growth hormone derivative.
Biochemistry (2020)
Johansson E, Nielsen AD, Demuth H, Wiberg C, Schjødt C, Huang T, Chen J, Jensen S, Petersen J, Thygesen P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
|
|
|
|
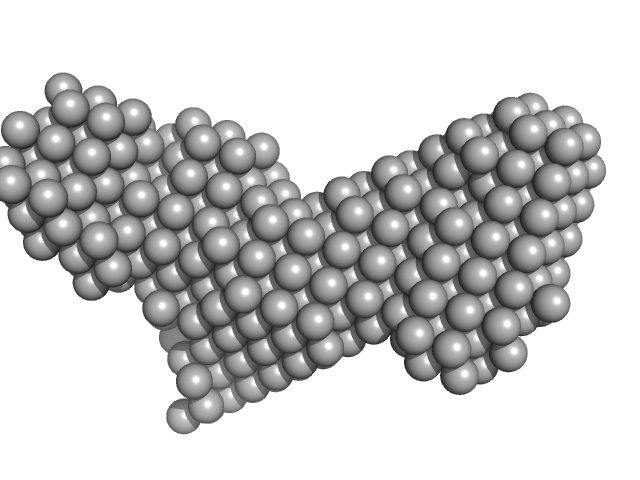
|
| Sample: |
Histone deacetylase 1 monomer, 55 kDa Homo sapiens protein
Lysine-specific histone demethylase 1A monomer, 93 kDa Homo sapiens protein
REST corepressor 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris/Cl, 50 mM potassium acetate and 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Jan 23
|
Mechanism of Crosstalk between the LSD1 Demethylase and HDAC1 Deacetylase in the CoREST Complex.
Cell Rep 30(8):2699-2711.e8 (2020)
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
|
| RgGuinier |
6.0 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
437 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Oct 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
147 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Palmitoyl-protein thioesterase 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
Palmitoyl-protein thioesterase 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
4.9 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
258 |
nm3 |
|
|
|
|
|
|
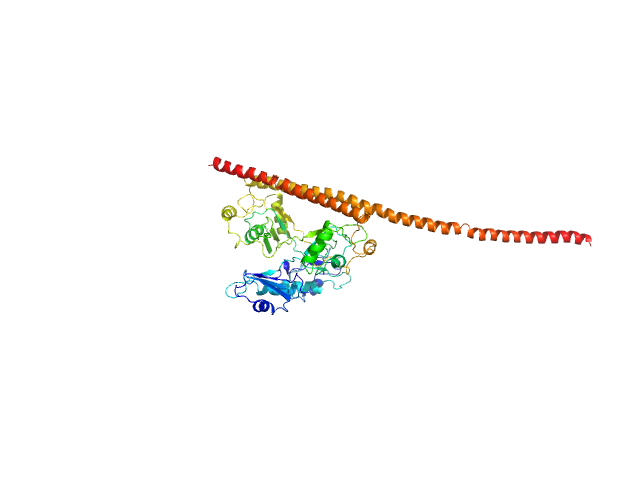
|
| Sample: |
Non-POU domain-containing octamer-binding protein monomer, 30 kDa Homo sapiens protein
Splicing factor, proline- and glutamine-rich monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 5
|
Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ
Nucleic Acids Research 48(6):3356-3365 (2020)
Huang J, Ringuet M, Whitten A, Caria S, Lim Y, Badhan R, Anggono V, Lee M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
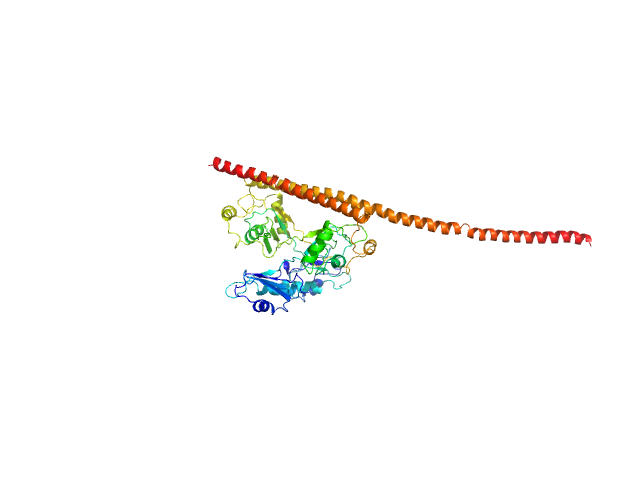
|
| Sample: |
Splicing factor, proline- and glutamine-rich monomer, 38 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 8
|
Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ
Nucleic Acids Research 48(6):3356-3365 (2020)
Huang J, Ringuet M, Whitten A, Caria S, Lim Y, Badhan R, Anggono V, Lee M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
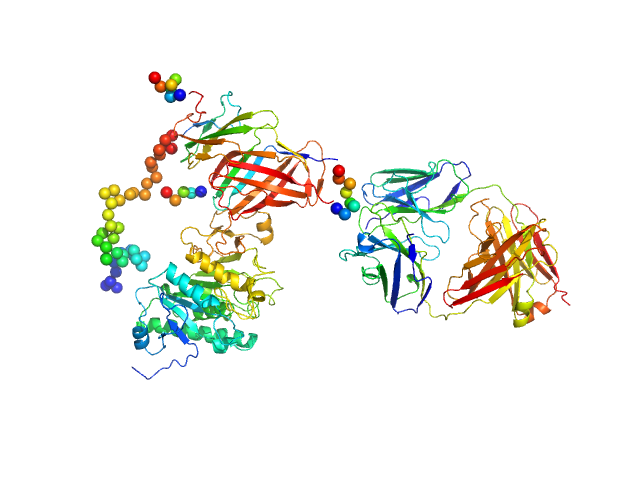
|
| Sample: |
Lipoprotein lipase monomer, 50 kDa Homo sapiens protein
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 monomer, 15 kDa Homo sapiens protein
Monoclonal Antibody Fragment 5D2 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, 4 mM CaCL2, 10% (v/v) Glycerol, 0.05% 0.8mM CHAPS, ,0.05 % (v/v) NaN3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 14
|
Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism
Proceedings of the National Academy of Sciences :201920202 (2020)
Kristensen K, Leth-Espensen K, Mertens H, Birrane G, Meiyappan M, Olivecrona G, Jørgensen T, Young S, Ploug M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
187 |
nm3 |
|
|