|
|
|
|
|
| Sample: |
Latency associated peptide dimer, 58 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Apr 20
|
Structural insights into conformational switching in latency-associated peptide between transforming growth factor β-1 bound and unbound states
IUCrJ 7(2) (2020)
Stachowski T, Snell M, Snell E
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Calcium-activated chloride channel regulator 1 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jun 6
|
Structural and Biophysical Analysis of the CLCA1 VWA Domain Suggests Mode of TMEM16A Engagement.
Cell Rep 30(4):1141-1151.e3 (2020)
Berry KN, Brett TJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
Calcium-activated chloride channel regulator 1 monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 6
|
Structural and Biophysical Analysis of the CLCA1 VWA Domain Suggests Mode of TMEM16A Engagement.
Cell Rep 30(4):1141-1151.e3 (2020)
Berry KN, Brett TJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.4 |
nm |
|
|
|
|
|
|
|
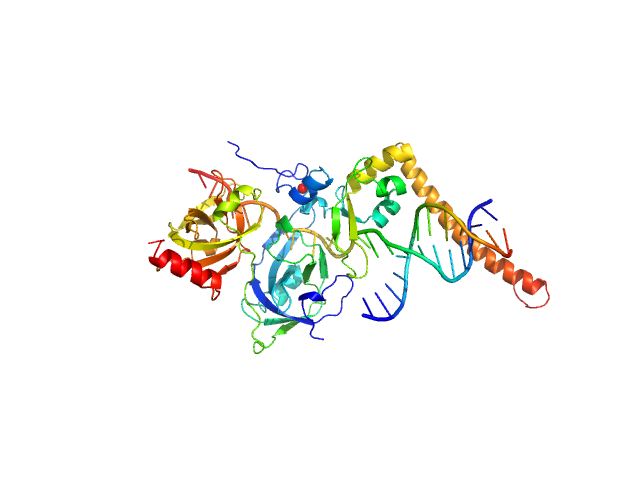
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
3-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 17 Nov 2
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
Topolska-Woś AM, Sugitani N, Cordoba JJ, Le Meur KV, Le Meur RA, Kim HS, Yeo JE, Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
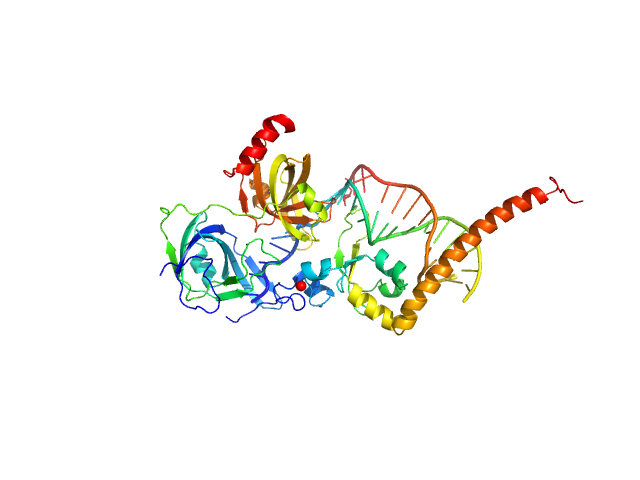
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
5-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 4
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
Topolska-Woś AM, Sugitani N, Cordoba JJ, Le Meur KV, Le Meur RA, Kim HS, Yeo JE, Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
2.9 |
nm |
| Dmax |
97.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
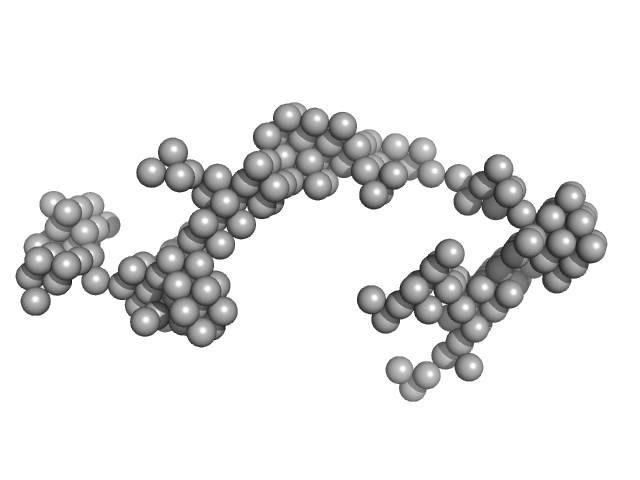
| Sample: |
Braveheart Fragment 1 monomer, 116 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
8.4 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
426 |
nm3 |
|
|
|
|
|
|
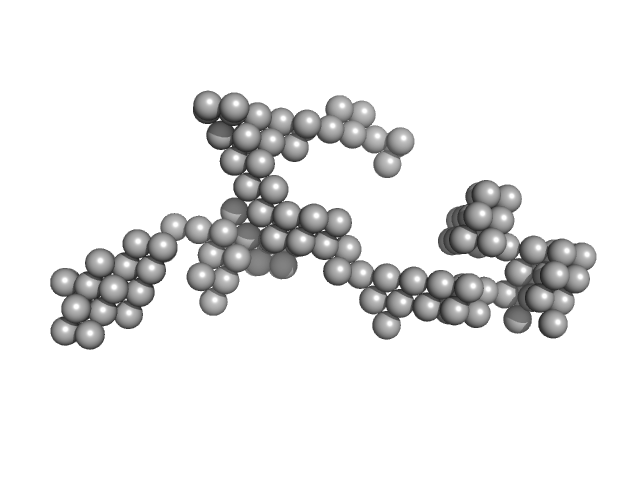
|
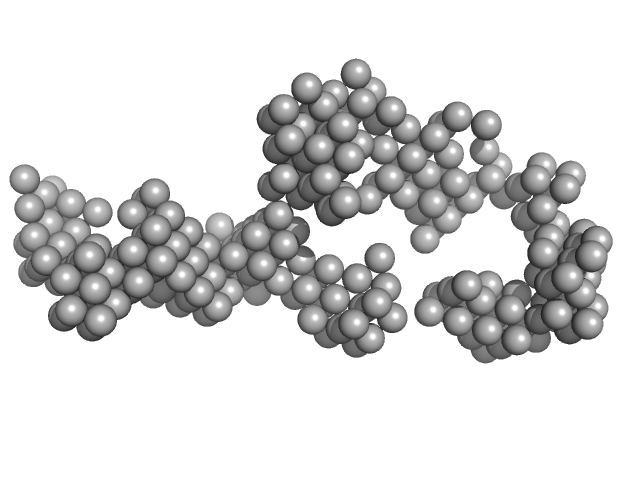
| Sample: |
Braveheart Fragment 2 monomer, 112 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
8.1 |
nm |
| Dmax |
27.2 |
nm |
| VolumePorod |
443 |
nm3 |
|
|
|
|
|
|
|
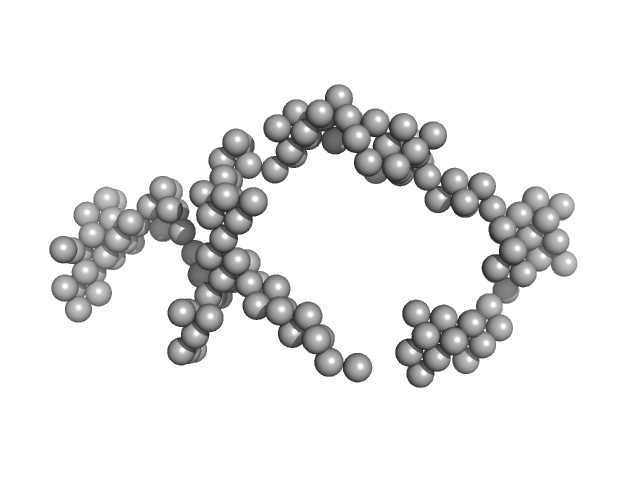
| Sample: |
Braveheart Fragment 3 monomer, 111 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
6.5 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
492 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
9.9 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
2150 |
nm3 |
|
|
|
|
|
|
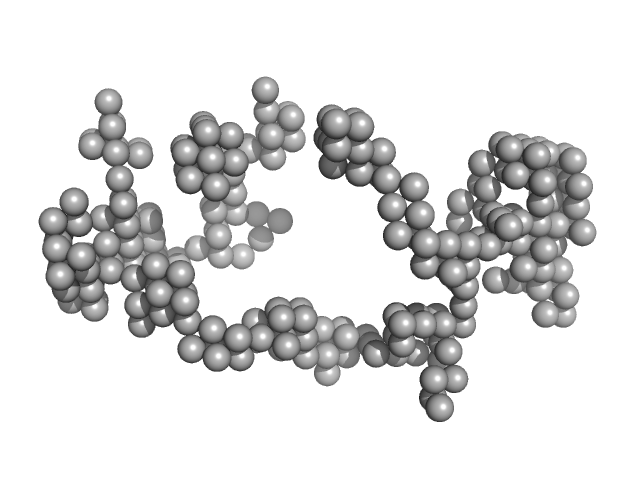
|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
11.8 |
nm |
| Dmax |
28.7 |
nm |
| VolumePorod |
2380 |
nm3 |
|
|