|
|
|
|
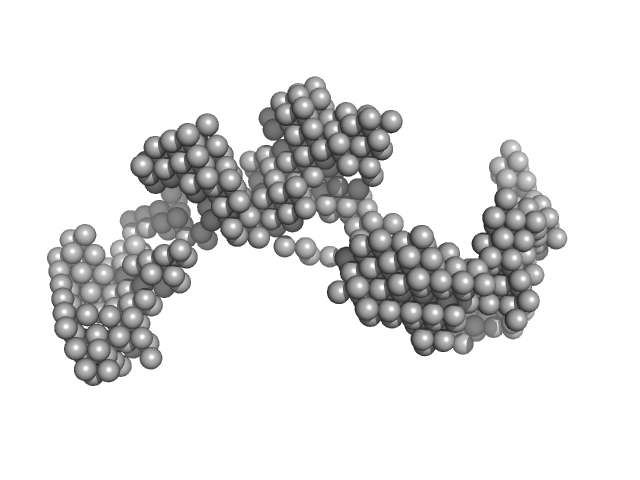
|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 12 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jul 20
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
9.7 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
1370 |
nm3 |
|
|
|
|
|
|
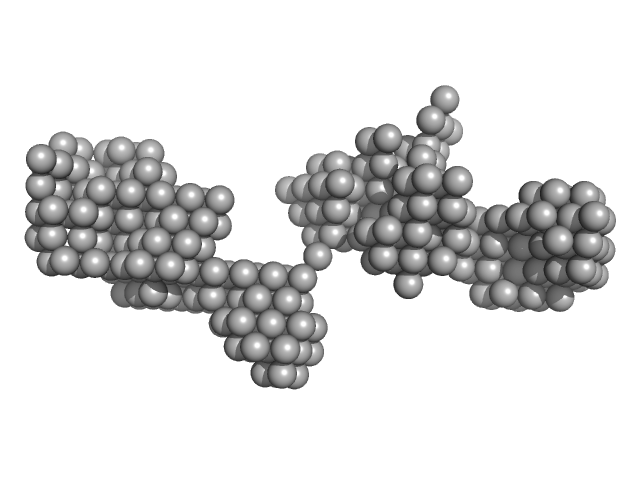
|
| Sample: |
Braveheart 5' Module monomer, 32 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
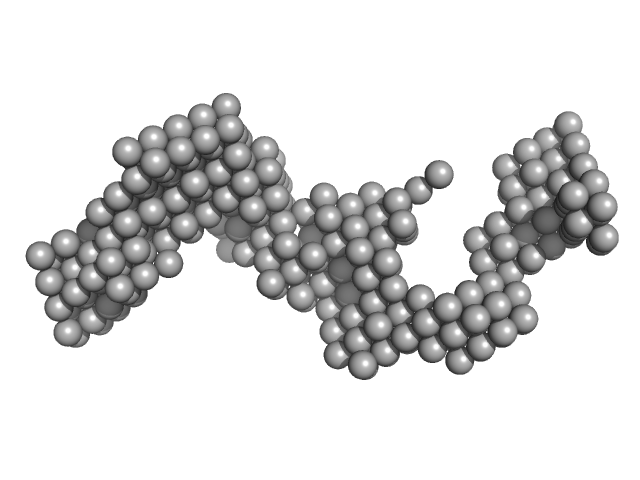
|
| Sample: |
Braveheart 5' Module monomer, 32 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 12 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
4.8 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
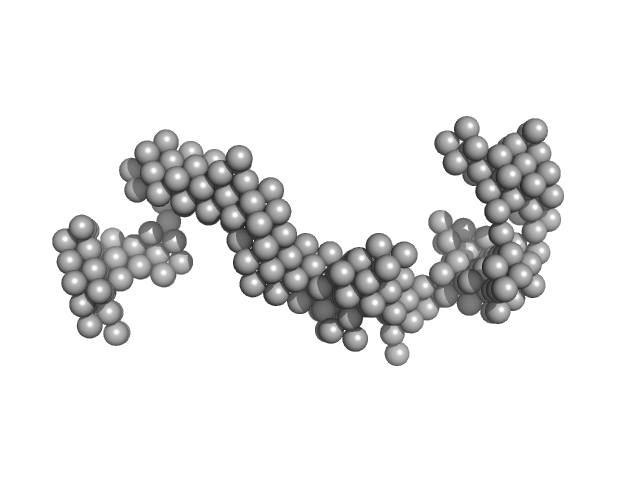
|
| Sample: |
Braveheart 3' module monomer, 72 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
6.5 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart 3' module monomer, 72 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 12 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
7.3 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
|
|
|
|
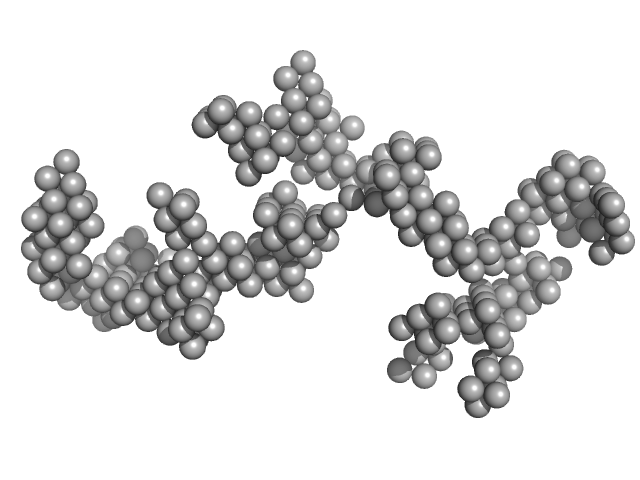
|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
9.8 |
nm |
| Dmax |
30.2 |
nm |
| VolumePorod |
1660 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
Braveheart Fragment 1 monomer, 116 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
8.2 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
455 |
nm3 |
|
|
|
|
|
|
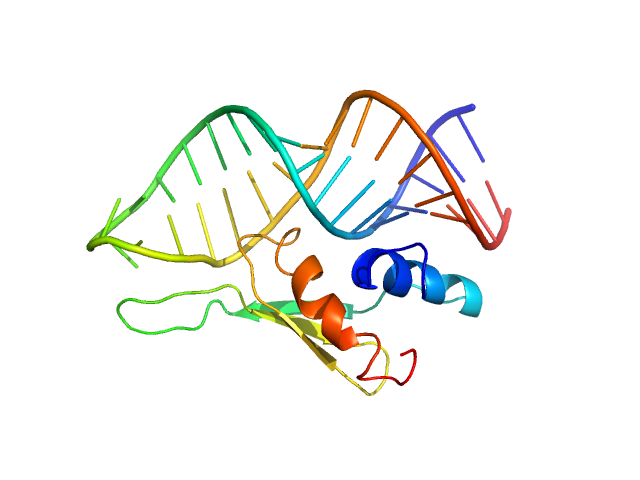
|
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 - RNA binding domain 4 monomer, 8 kDa Homo sapiens protein
ADP-ribosylation factor1 - short monomer, 11 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 3.5 mM 2-mercaptoethanol, pH: 6.8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2015 Dec 10
|
Staufen1 reads out structure and sequence features in ARF1 dsRNA for target recognition.
Nucleic Acids Res (2019)
Yadav DK, Zigáčková D, Zlobina M, Klumpler T, Beaumont C, Kubíčková M, Vaňáčová Š, Lukavsky PJ
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
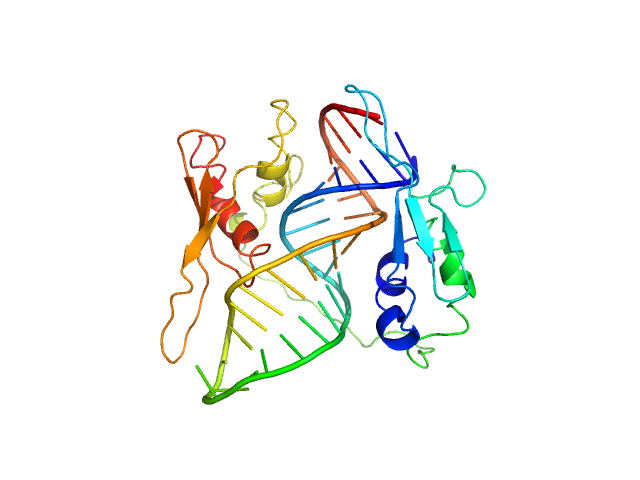
|
| Sample: |
ADP-ribosylation factor1 - short monomer, 11 kDa Homo sapiens RNA
Double-stranded RNA-binding protein Staufen homolog 1 - RNA binding domain 3 and 4 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, 300 mM NaCl, 500 mM imidazole, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2018 May 29
|
Staufen1 reads out structure and sequence features in ARF1 dsRNA for target recognition.
Nucleic Acids Res (2019)
Yadav DK, Zigáčková D, Zlobina M, Klumpler T, Beaumont C, Kubíčková M, Vaňáčová Š, Lukavsky PJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 - RNA binding domain 3 and 4 monomer, 20 kDa Homo sapiens protein
ADP-ribosylation factor1 - long monomer, 16 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 3.5 mM 2-mercaptoethanol, pH: 6.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 20
|
Staufen1 reads out structure and sequence features in ARF1 dsRNA for target recognition.
Nucleic Acids Res (2019)
Yadav DK, Zigáčková D, Zlobina M, Klumpler T, Beaumont C, Kubíčková M, Vaňáčová Š, Lukavsky PJ
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|