|
|
|
|
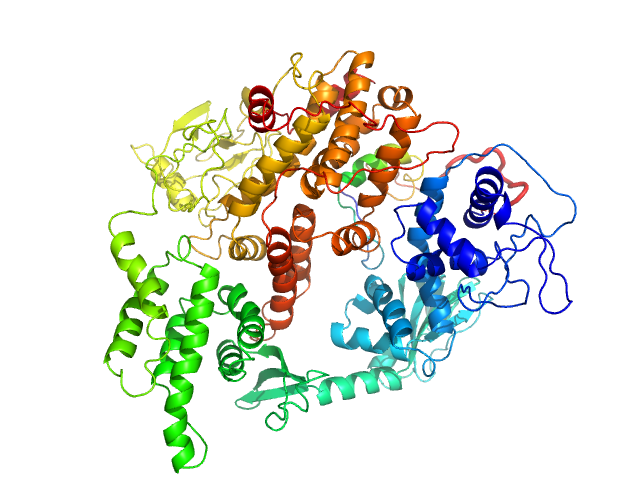
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2012 Sep 7
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
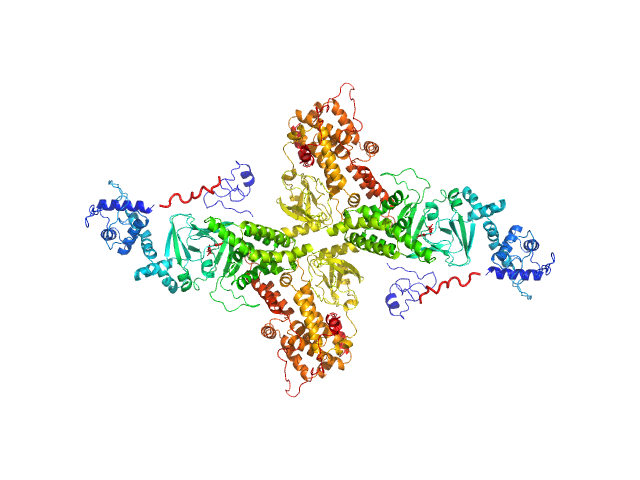
|
| Sample: |
Rap guanine nucleotide exchange factor 3 (dimer) dimer, 200 kDa Homo sapiens protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, 1mM cAMP, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2012 Jan 30
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
5.3 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
415 |
nm3 |
|
|
|
|
|
|
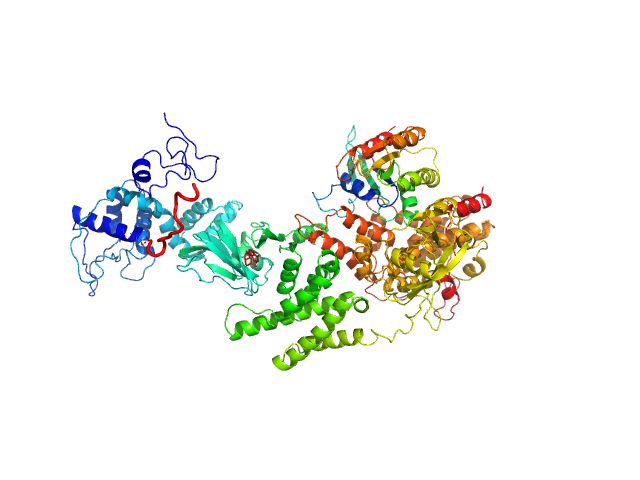
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 100 kDa Homo sapiens protein
RAS related protein 1b monomer, 18 kDa Mus musculus protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, 1mM cAMP, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2013 Apr 1
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|
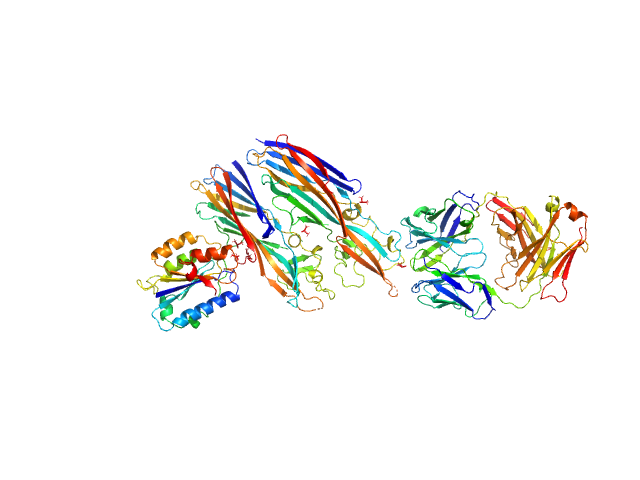
|
| Sample: |
Leukocidin G monomer, 36 kDa Staphylococcus aureus protein
Leukocidin H monomer, 38 kDa Staphylococcus aureus protein
LukGH neutralizing antibody monomer, 47 kDa Homo sapiens protein
Integrin alpha-M monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 17
|
Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity.
Proc Natl Acad Sci U S A (2019)
Trstenjak N, Milić D, Graewert MA, Rouha H, Svergun D, Djinović-Carugo K, Nagy E, Badarau A
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
178 |
nm3 |
|
|
|
|
|
|
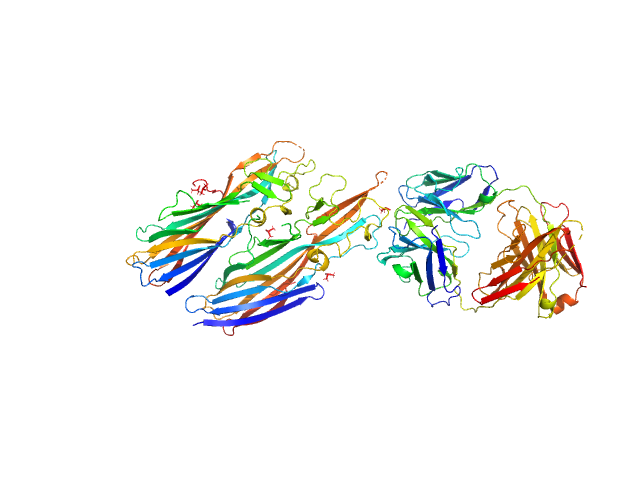
|
| Sample: |
Leukocidin G monomer, 36 kDa Staphylococcus aureus protein
Leukocidin H monomer, 38 kDa Staphylococcus aureus protein
LukGH neutralizing antibody monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 17
|
Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity.
Proc Natl Acad Sci U S A (2019)
Trstenjak N, Milić D, Graewert MA, Rouha H, Svergun D, Djinović-Carugo K, Nagy E, Badarau A
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
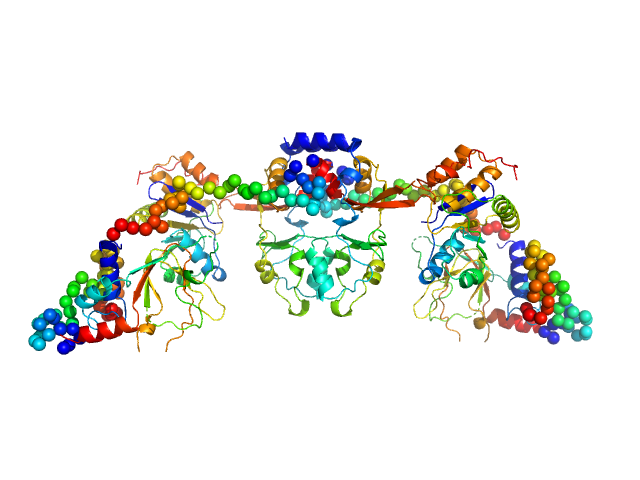
|
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
Protein tyrosine phosphatase type IVA 1 dimer, 40 kDa Mus musculus protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 17
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks T, Blanco FJ, Díaz Moreno I, Martínez-Chantar ML, Tremblay ML, Müller D, Siliqi D, Martínez-Cruz LA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
216 |
nm3 |
|
|
|
|
|
|
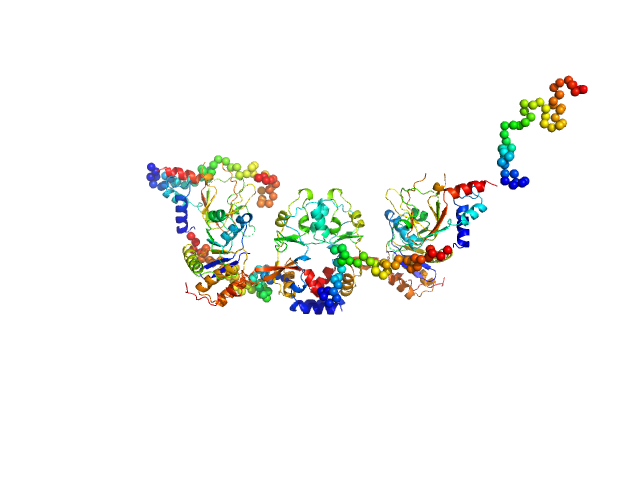
|
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
Protein tyrosine phosphatase type IVA 1 dimer, 40 kDa Mus musculus protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Aug 2
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks T, Blanco FJ, Díaz Moreno I, Martínez-Chantar ML, Tremblay ML, Müller D, Siliqi D, Martínez-Cruz LA
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
|
|
|
|
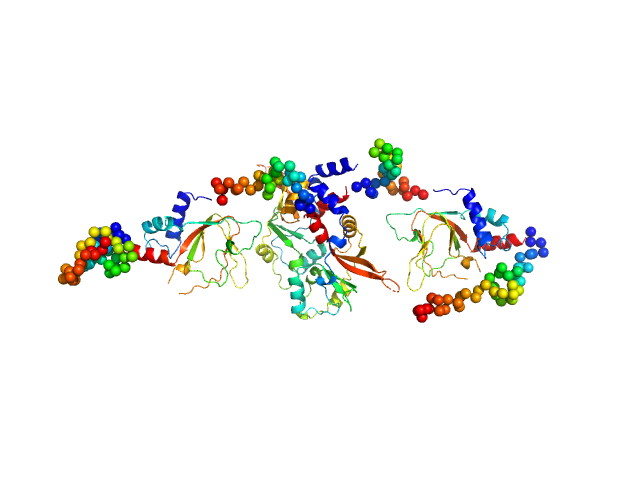
|
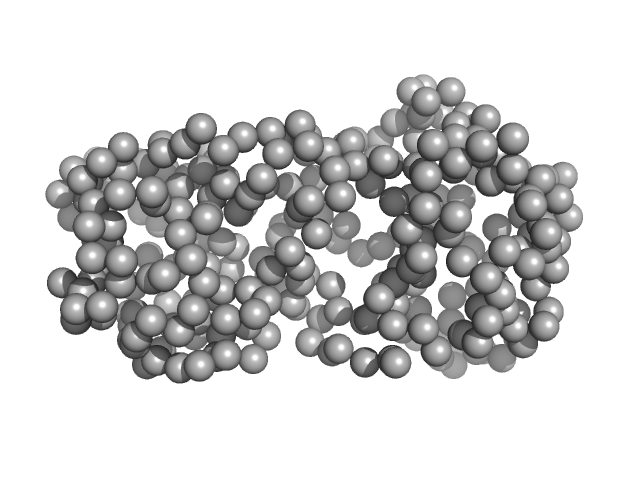
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Aug 2
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks T, Blanco FJ, Díaz Moreno I, Martínez-Chantar ML, Tremblay ML, Müller D, Siliqi D, Martínez-Cruz LA
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.6 |
nm |
| VolumePorod |
156 |
nm3 |
|
|
|
|
|
|
|
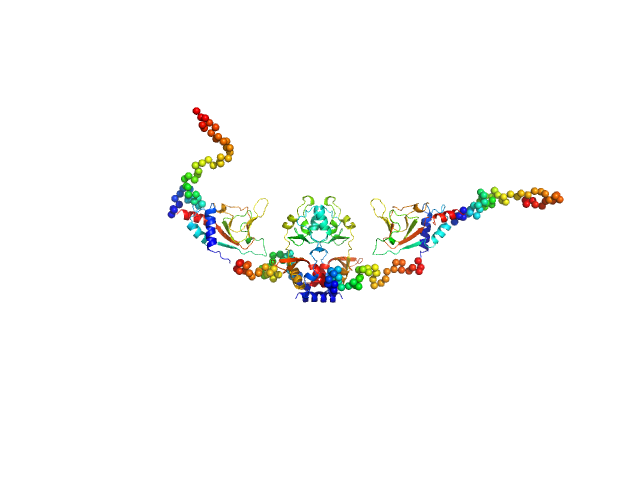
| Sample: |
CNNM4_cNMP dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 7
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks T, Blanco FJ, Díaz Moreno I, Martínez-Chantar ML, Tremblay ML, Müller D, Siliqi D, Martínez-Cruz LA
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 17
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks T, Blanco FJ, Díaz Moreno I, Martínez-Chantar ML, Tremblay ML, Müller D, Siliqi D, Martínez-Cruz LA
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
149 |
nm3 |
|
|