|
|
|
|
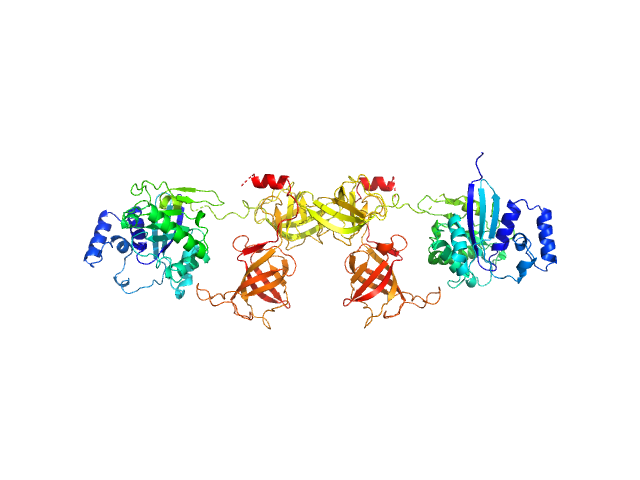
|
| Sample: |
Mammalian translation elongation factor eEF1A1 dimer, 100 kDa protein
|
| Buffer: |
25 mM Tris HCl, 150 mM NaCl, 6 mM βME, 20% glycerol, 0.01mM GDP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2023 Mar 20
|
Dynamics and structural features of the eEF1A1 and eEF1A2 paralogs
Nucleic Acids Research 53(21) (2025)
Novosylna O, Shalak V, Dąbrowska K, Patmanidis I, Lozhko D, Bondarchuk T, Schiøtt B, Pedersen J, Knudsen C, Nissen P, Dadlez M, Negrutskii B
|
| RgGuinier |
4.6 |
nm |
| Dmax |
143.1 |
nm |
| VolumePorod |
144 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hen egg white lysozyme monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Jul 9
|
Lysozyme pH 7.4 with models
Anthony Giannetti
|
| RgGuinier |
1.6 |
nm |
| Dmax |
59.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
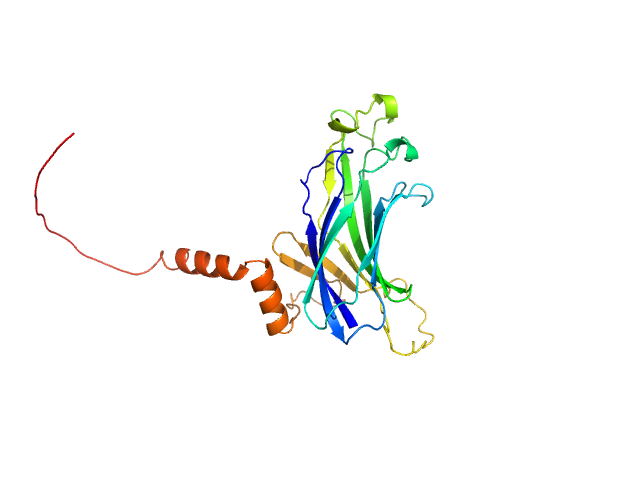
|
| Sample: |
Anti-Silencing Function 1 monomer, 28 kDa Entamoeba histolytica strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, and 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Oct 1
|
Characterisation of Entamoeba histolytica Anti-Silencing Function 1 as a histone chaperone.
Biochimie (2025)
Gandhi S, Vasudevan D
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Anti-Silencing Function 1 monomer, 28 kDa Entamoeba histolytica strain … protein
Histone H3 monomer, 15 kDa Entamoeba histolytica strain … protein
Histone H4 monomer, 13 kDa Entamoeba histolytica strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, and 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Oct 1
|
Characterisation of Entamoeba histolytica Anti-Silencing Function 1 as a histone chaperone.
Biochimie (2025)
Gandhi S, Vasudevan D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
N-acetylmuramoyl-L-alanine amidase monomer, 43 kDa Mycobacterium phage DS6A protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl and 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Sep 30
|
Dissecting the molecular basis underlying mycobacterial cell-wall hydrolysis by the catalytic domains of D29LysA and DS6ALysA phage endolysins.
Int J Biol Macromol :148896 (2025)
Ceballos-Zúñiga F, Galvez-Larrosa L, Muñoz IG, Infantes L, Fernández-Carrillo J, Pérez-Dorado I
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
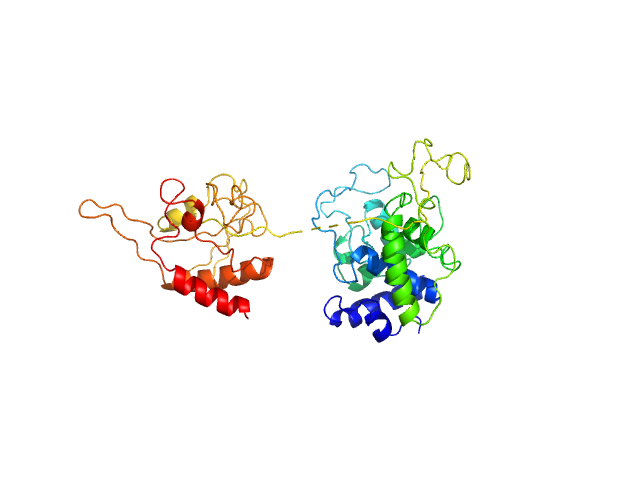
|
| Sample: |
N-acetylmuramoyl-L-alanine amidase (Mycobacteriophage DS6A) monomer, 45 kDa protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl and 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 30
|
Dissecting the molecular basis underlying mycobacterial cell-wall hydrolysis by the catalytic domains of D29LysA and DS6ALysA phage endolysins.
Int J Biol Macromol :148896 (2025)
Ceballos-Zúñiga F, Galvez-Larrosa L, Muñoz IG, Infantes L, Fernández-Carrillo J, Pérez-Dorado I
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
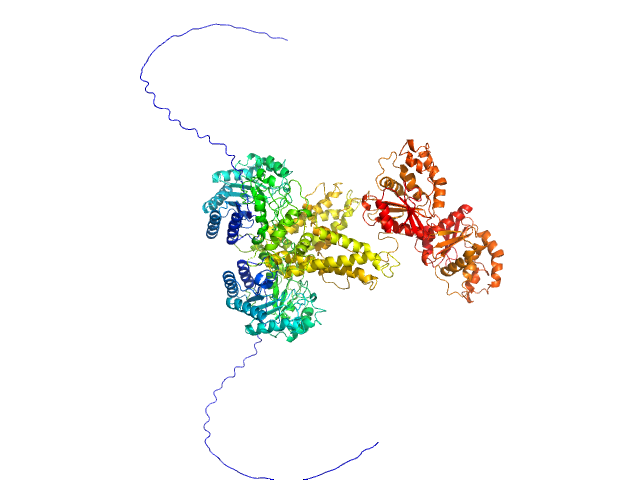
|
| Sample: |
RTX cytotoxin pro-MbxA without acylation monomer, 100 kDa Moraxella bovis protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 10mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 9
|
Acylation of the RTX toxin MbxA stimulates host membrane disruption through a specific interaction with cholesterol
Biochimica et Biophysica Acta (BBA) - Biomembranes :184487 (2025)
Chacko F, Ganz S, Pfitzer-Bilsing A, Hänsch S, Westhoff P, Weidtkamp-Peters S, Smits S, Exterkate M, Schmitt L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|
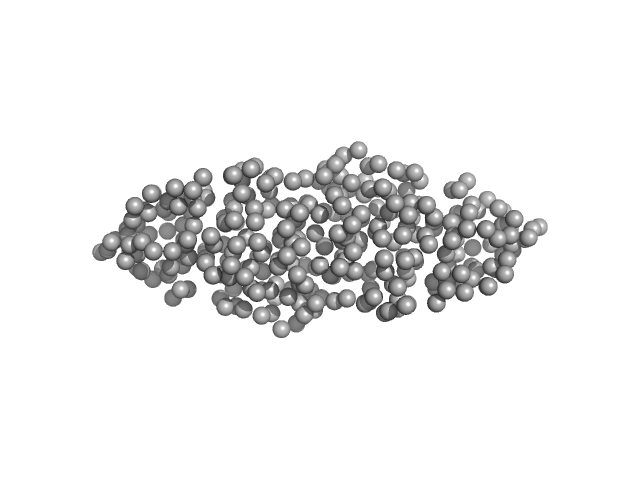
|
| Sample: |
Mutual gliding motility protein C dimer, 32 kDa Myxococcus xanthus (strain … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 31
|
Structural and biophysical insights into RomR, MglB and MglC interactions involved in regulating cell polarity in Myxococcus xanthus
Journal of Biological Chemistry :110907 (2025)
Kodesia A, Kapoor S, Thakur K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
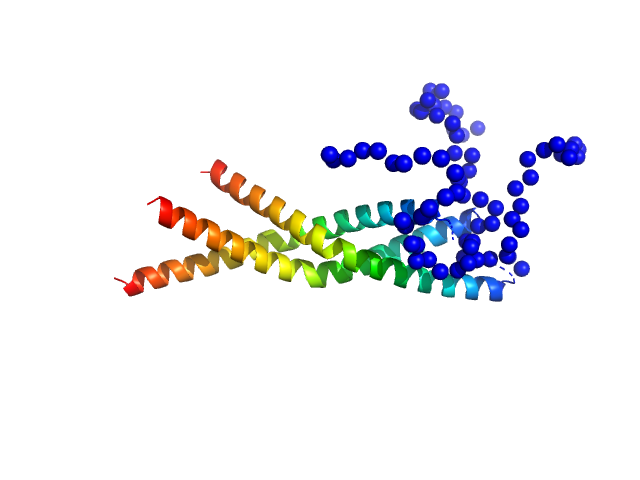
|
| Sample: |
Two-component system response regulator trimer, 26 kDa Myxococcus xanthus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 31
|
Structural and biophysical insights into RomR, MglB and MglC interactions involved in regulating cell polarity in Myxococcus xanthus
Journal of Biological Chemistry :110907 (2025)
Kodesia A, Kapoor S, Thakur K
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Two-component system response regulator trimer, 39 kDa Myxococcus xanthus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 31
|
Structural and biophysical insights into RomR, MglB and MglC interactions involved in regulating cell polarity in Myxococcus xanthus
Journal of Biological Chemistry :110907 (2025)
Kodesia A, Kapoor S, Thakur K
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
81 |
nm3 |
|
|