|
|
|
|
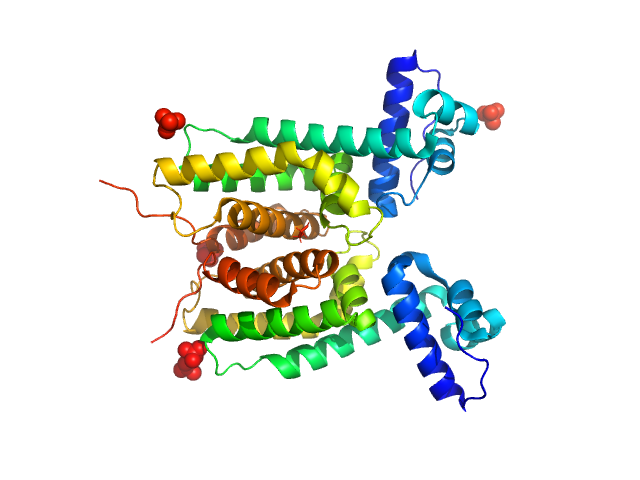
|
| Sample: |
LuxR family transcriptional regulator, N55I, 52 kDa Vibrio vulnificus protein
|
| Buffer: |
200 mM NaCl, 25 mM Tris, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 11
|
The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res 49(10):5967-5984 (2021)
Newman JD, Russell MM, Fan L, Wang YX, Gonzalez-Gutierrez G, van Kessel JC
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
M18D170V209D170M18 monomer, 1 kDa
|
| Buffer: |
H20, pH: 2.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 5
|
Highly Tunable Nanostructures in a Doubly pH‐Responsive Pentablock Terpolymer in Solution and in Thin Films
Advanced Functional Materials 31(32):2102905 (2021)
Jung F, Schart M, Bührend L, Meidinger E, Kang J, Niebuur B, Ariaee S, Molodenskiy D, Posselt D, Amenitsch H, Tsitsilianis C, Papadakis C
|
|
|
|
|
|
|
|
| Sample: |
M18D170V209D170M18 monomer, 1 kDa
|
| Buffer: |
H2O, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 5
|
Highly Tunable Nanostructures in a Doubly pH‐Responsive Pentablock Terpolymer in Solution and in Thin Films
Advanced Functional Materials 31(32):2102905 (2021)
Jung F, Schart M, Bührend L, Meidinger E, Kang J, Niebuur B, Ariaee S, Molodenskiy D, Posselt D, Amenitsch H, Tsitsilianis C, Papadakis C
|
|
|
|
|
|
|
|
| Sample: |
M18D170V209D170M18 monomer, 1 kDa
|
| Buffer: |
H2O, pH: 6.9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 5
|
Highly Tunable Nanostructures in a Doubly pH‐Responsive Pentablock Terpolymer in Solution and in Thin Films
Advanced Functional Materials 31(32):2102905 (2021)
Jung F, Schart M, Bührend L, Meidinger E, Kang J, Niebuur B, Ariaee S, Molodenskiy D, Posselt D, Amenitsch H, Tsitsilianis C, Papadakis C
|
|
|
|
|
|
|
|
| Sample: |
M18D170V209D170M18 monomer, 1 kDa
|
| Buffer: |
H2O, pH: 8.9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 5
|
Highly Tunable Nanostructures in a Doubly pH‐Responsive Pentablock Terpolymer in Solution and in Thin Films
Advanced Functional Materials 31(32):2102905 (2021)
Jung F, Schart M, Bührend L, Meidinger E, Kang J, Niebuur B, Ariaee S, Molodenskiy D, Posselt D, Amenitsch H, Tsitsilianis C, Papadakis C
|
|
|
|
|
|
|

|
| Sample: |
Hybrid RTX-1 construct (amino acids 1132-1294 and 1562-1681 of CyaA) monomer, 30 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 1
|
Almost half of the RTX domain is dispensable for complement receptor 3 binding and cell-invasive activity of the adenylate cyclase toxin.
J Biol Chem :100833 (2021)
Espinosa-Vinals CA, Masin J, Holubova J, Stanek O, Jurnecka D, Osicka R, Sebo P, Bumba L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hybrid RTX-2 construct (amino acids 1132-1303 and 1562-1681 of CyaA) monomer, 31 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 1
|
Almost half of the RTX domain is dispensable for complement receptor 3 binding and cell-invasive activity of the adenylate cyclase toxin.
J Biol Chem :100833 (2021)
Espinosa-Vinals CA, Masin J, Holubova J, Stanek O, Jurnecka D, Osicka R, Sebo P, Bumba L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
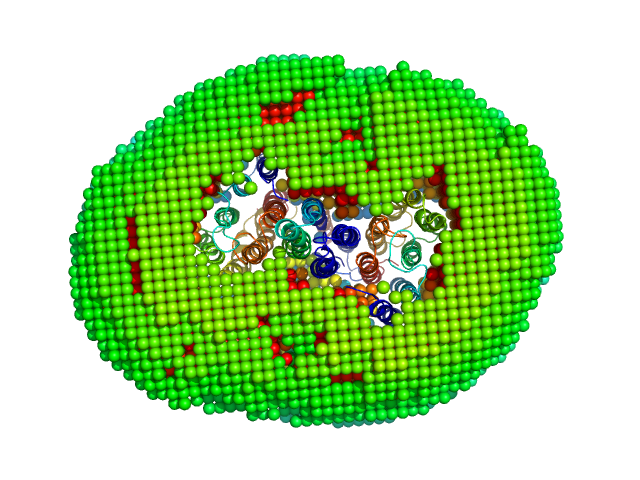
|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Truncated sensory rhodopsin II transducer containing transmembrane and HAMP1 domains dimer, 30 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 Na/K-Pi, 1 mM EDTA, 0.05 % DDM, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 13
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 9
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
9.0 |
nm |
| Dmax |
38.9 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
7.1 |
nm |
| Dmax |
35.0 |
nm |
|
|