|
|
|
|
|
| Sample: |
Salt stress-induced protein dimer, 37 kDa Oryza sativa Indica … protein
|
| Buffer: |
50 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2017 Mar 6
|
Structural insights into rice SalTol QTL located SALT protein.
Sci Rep 10(1):16589 (2020)
Kaur N, Sagar A, Sharma P, Ashish, Pati PK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 16 kDa Gallus gallus protein
|
| Buffer: |
40 mM sodium acetate, 150 mM NaCl, 100 mM 5-methyl uridine, pH: 3.8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Jun 7
|
Improving data quality and expanding BioSAXS experiments to low-molecular-weight and low-concentration protein samples.
Acta Crystallogr D Struct Biol 76(Pt 10):971-981 (2020)
Castellví A, Pascual-Izarra C, Crosas E, Malfois M, Juanhuix J
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.2 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Spectrin alpha chain, non-erythrocytic 1, 7 kDa Gallus gallus protein
|
| Buffer: |
10 mM citric acid, 150 mM NaCl, 100 mM 5-methyl uridine, pH: 3 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Jun 14
|
Improving data quality and expanding BioSAXS experiments to low-molecular-weight and low-concentration protein samples.
Acta Crystallogr D Struct Biol 76(Pt 10):971-981 (2020)
Castellví A, Pascual-Izarra C, Crosas E, Malfois M, Juanhuix J
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HSP40, subfamily A dimer, 97 kDa Plasmodium falciparum protein
|
| Buffer: |
50 mM Tris, 5 mM MgCl2, 300 mM KCl, 1 mM β-mercaptoethanol, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Oct 24
|
Structural-functional diversity of malaria parasite's PfHSP70-1 and PfHSP40 chaperone pair gives an edge over human orthologs in chaperone-assisted protein folding.
Biochem J 477(18):3625-3643 (2020)
Anas M, Shukla A, Tripathi A, Kumari V, Prakash C, Nag P, Kumar LS, Sharma SK, Ramachandran R, Kumar N
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
339 |
nm3 |
|
|
|
|
|
|
|
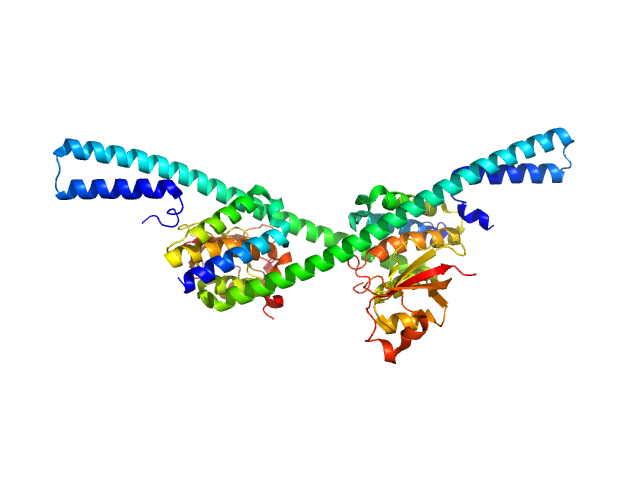
| Sample: |
Replicase polyprotein 1a - nsp7, 18 kDa Severe acute respiratory … protein
Replicase polyprotein 1a - nsp8, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, pH 8.0, 100 mM NaCl, 4 mM DTT, 4 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 19
|
Hallmarks of Alpha-
and Betacoronavirus
non-structural protein 7+8 complexes
Science Advances 7(10):eabf1004 (2021)
Krichel B, Bylapudi G, Schmidt C, Blanchet C, Schubert R, Brings L, Koehler M, Zenobi R, Svergun D, Lorenzen K, Madhugiri R, Ziebuhr J, Uetrecht C
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Photinus pyralis firefly luciferase, 61 kDa Photinus pyralis protein
|
| Buffer: |
TBS (20 mM Tris, 150 mM NaCl),, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 3
|
Rapid screening of in cellulo
grown protein crystals via a small-angle X-ray scattering/X-ray powder diffraction synergistic approach
Journal of Applied Crystallography 53(5) (2020)
Lahey-Rudolph J, Schönherr R, Jeffries C, Blanchet C, Boger J, Ferreira Ramos A, Riekehr W, Triandafillidis D, Valmas A, Margiolaki I, Svergun D, Redecke L
|
|
|
|
|
|
|
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase, 59 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
TBS (20 mM Tris, 150 mM NaCl),, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 3
|
Rapid screening of in cellulo
grown protein crystals via a small-angle X-ray scattering/X-ray powder diffraction synergistic approach
Journal of Applied Crystallography 53(5) (2020)
Lahey-Rudolph J, Schönherr R, Jeffries C, Blanchet C, Boger J, Ferreira Ramos A, Riekehr W, Triandafillidis D, Valmas A, Margiolaki I, Svergun D, Redecke L
|
|
|
|
|
|
|
|
| Sample: |
Cathepsin B-like cysteine protease, 37 kDa Trypanosoma brucei protein
|
| Buffer: |
TBS (20 mM Tris, 150 mM NaCl),, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 3
|
Rapid screening of in cellulo
grown protein crystals via a small-angle X-ray scattering/X-ray powder diffraction synergistic approach
Journal of Applied Crystallography 53(5) (2020)
Lahey-Rudolph J, Schönherr R, Jeffries C, Blanchet C, Boger J, Ferreira Ramos A, Riekehr W, Triandafillidis D, Valmas A, Margiolaki I, Svergun D, Redecke L
|
|
|
|
|
|
|
|
| Sample: |
Woronin body major protein, 19 kDa Neurospora crassa protein
|
| Buffer: |
TBS (20 mM Tris, 150 mM NaCl),, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 3
|
Rapid screening of in cellulo
grown protein crystals via a small-angle X-ray scattering/X-ray powder diffraction synergistic approach
Journal of Applied Crystallography 53(5) (2020)
Lahey-Rudolph J, Schönherr R, Jeffries C, Blanchet C, Boger J, Ferreira Ramos A, Riekehr W, Triandafillidis D, Valmas A, Margiolaki I, Svergun D, Redecke L
|
|
|
|
|
|
|
|
| Sample: |
Mock recombinant baculovirus, 0 kDa unidentified baculovirus
|
| Buffer: |
TBS (20 mM Tris, 150 mM NaCl),, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 3
|
Rapid screening of in cellulo
grown protein crystals via a small-angle X-ray scattering/X-ray powder diffraction synergistic approach
Journal of Applied Crystallography 53(5) (2020)
Lahey-Rudolph J, Schönherr R, Jeffries C, Blanchet C, Boger J, Ferreira Ramos A, Riekehr W, Triandafillidis D, Valmas A, Margiolaki I, Svergun D, Redecke L
|
|
|