|
|
|
|
|
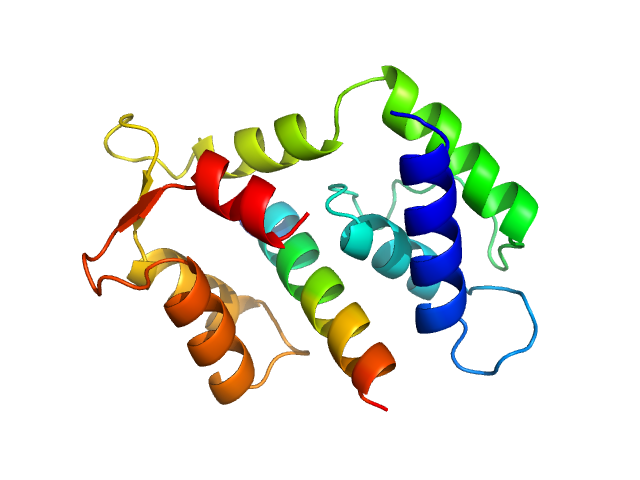
| Sample: |
Myosin essential light chain 2 monomer, 15 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myosin essential light chain 2 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 3 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
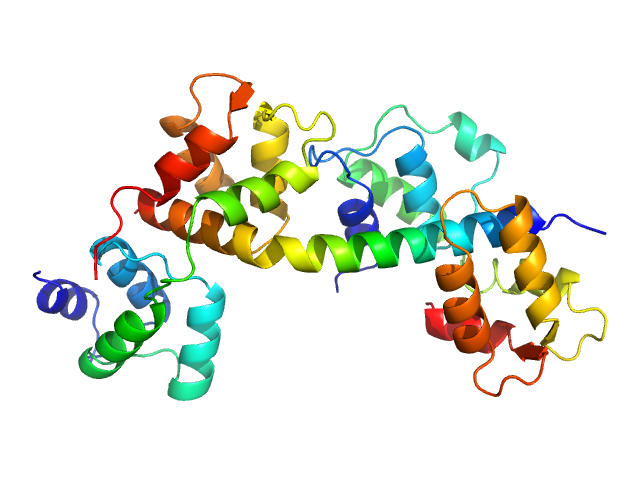
| Sample: |
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
Toxoplasma gondii myosin light chain, full length monomer, 24 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
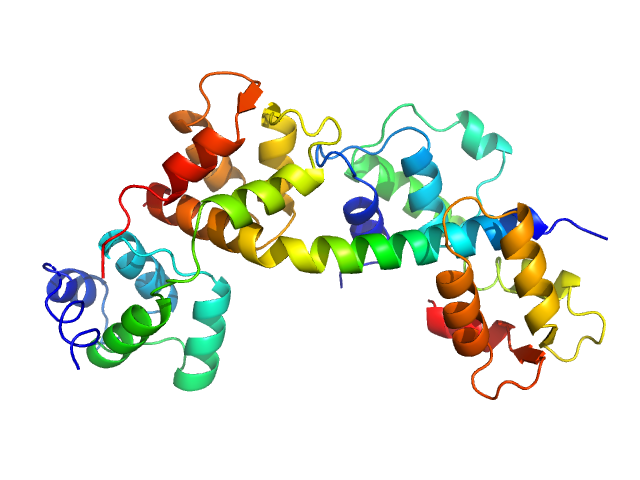
| Sample: |
Myosin light chain TgMLC1, residues 66-210 monomer, 17 kDa Toxoplasma gondii protein
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
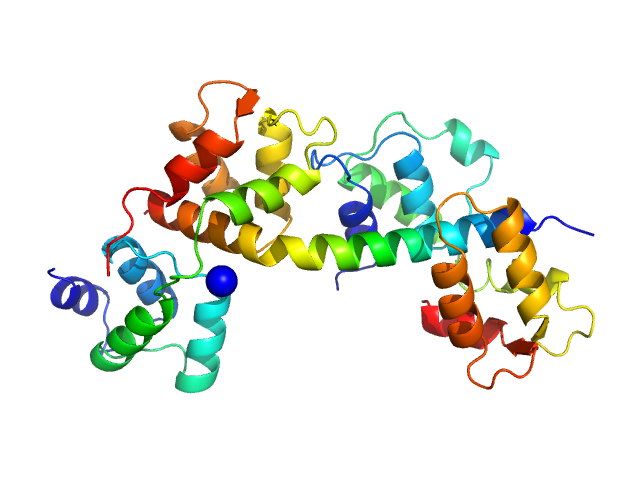
| Sample: |
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
Toxoplasma gondii myosin light chain, residues 70-210 monomer, 17 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
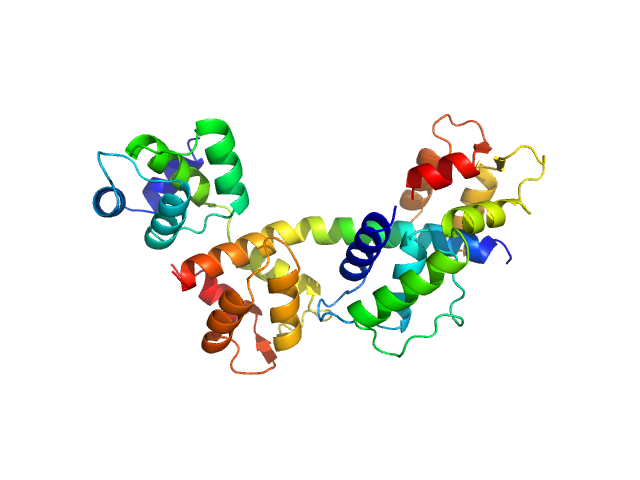
| Sample: |
Myosin essential light chain 2 monomer, 15 kDa Toxoplasma gondii protein
Myosin light chain TgMLC1, residues 66-210 monomer, 17 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myosin essential light chain monomer, 16 kDa Plasmodium falciparum protein
Plasmodium falciparum myosin A monomer, 5 kDa Plasmodium falciparum protein
Myosin A tail domain interacting protein monomer, 17 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ataxin-3 (polyglutamine protein ataxin-3 (Q54)) monomer, 46 kDa protein
|
| Buffer: |
20 mM sodium phosphate buffer, 2 mM TCEP, 5% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 24
|
Capturing the Conformational Ensemble of the Mixed Folded Polyglutamine Protein Ataxin-3.
Structure (2020)
Sicorello A, Różycki B, Konarev PV, Svergun DI, Pastore A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Salt stress-induced protein dimer, 37 kDa Oryza sativa Indica … protein
|
| Buffer: |
50 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2017 Mar 6
|
Structural insights into rice SalTol QTL located SALT protein.
Sci Rep 10(1):16589 (2020)
Kaur N, Sagar A, Sharma P, Ashish, Pati PK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Salt stress-induced protein dimer, 37 kDa Oryza sativa Indica … protein
|
| Buffer: |
50 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2017 Mar 6
|
Structural insights into rice SalTol QTL located SALT protein.
Sci Rep 10(1):16589 (2020)
Kaur N, Sagar A, Sharma P, Ashish, Pati PK
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
60 |
nm3 |
|
|