|
|
|
|

|
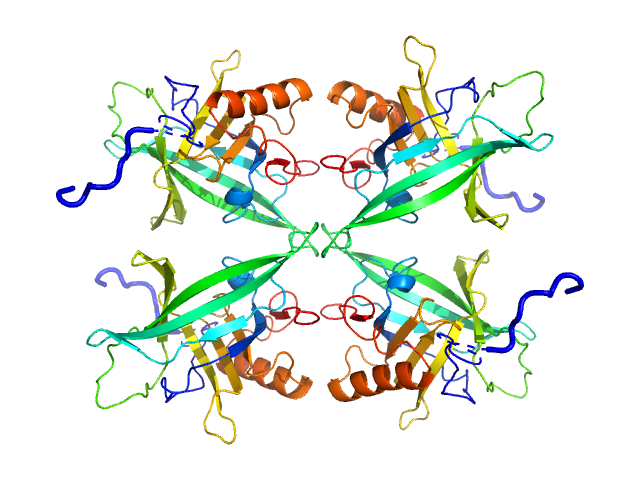
| Sample: |
Aquifex aeolicus McoA metaloxidase monomer, 55 kDa Aquifex aeolicus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 15
|
The Methionine-Rich Loop of Multicopper Oxidase McoA follows Open-To-Close Transitions with a Role in Enzyme Catalysis
ACS Catalysis (2020)
Borges P, Brissos V, Hernandez G, Masgrau L, Lucas M, Monza E, Frazão C, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
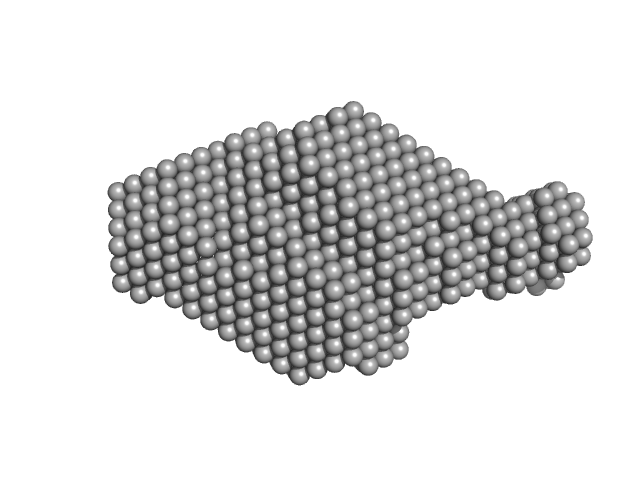
| Sample: |
Plasmodium falciparum Lipocalin tetramer, 89 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM Tris pH7.5, 150 mM NaCl, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum
Cell Reports 31(12):107817 (2020)
Burda P, Crosskey T, Lauk K, Zurborg A, Söhnchen C, Liffner B, Wilcke L, Pietsch E, Strauss J, Jeffries C, Svergun D, Wilson D, Wilmanns M, Gilberger T
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tegument protein UL7 monomer, 34 kDa Human alphaherpesvirus 1 … protein
Tegument protein UL51 dimer, 30 kDa Human alphaherpesvirus 1 … protein
|
| Buffer: |
20 mM tris, 200 mM NaCl, 3% (v/v) glycerol, 0.25 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 16
|
Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex.
Elife 9 (2020)
Butt BG, Owen DJ, Jeffries CM, Ivanova L, Hill CH, Houghton JW, Ahmed MF, Antrobus R, Svergun DI, Welch JJ, Crump CM, Graham SC
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|

|
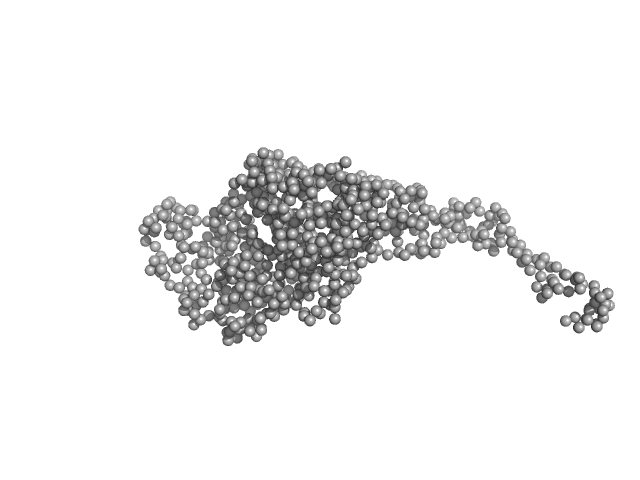
| Sample: |
Tegument protein UL7 dimer, 68 kDa Human alphaherpesvirus 1 … protein
Tegument protein UL51 tetramer, 102 kDa Human alphaherpesvirus 1 … protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 16
|
Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex.
Elife 9 (2020)
Butt BG, Owen DJ, Jeffries CM, Ivanova L, Hill CH, Houghton JW, Ahmed MF, Antrobus R, Svergun DI, Welch JJ, Crump CM, Graham SC
|
| RgGuinier |
4.6 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tegument protein UL7 monomer, 34 kDa Human alphaherpesvirus 1 … protein
Tegument protein UL51 dimer, 51 kDa Human alphaherpesvirus 1 protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 16
|
Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex.
Elife 9 (2020)
Butt BG, Owen DJ, Jeffries CM, Ivanova L, Hill CH, Houghton JW, Ahmed MF, Antrobus R, Svergun DI, Welch JJ, Crump CM, Graham SC
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.2 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cytoplasmic envelopment protein 3 monomer, 12 kDa Human alphaherpesvirus 1 protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP),, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 4
|
Conserved Outer Tegument Component UL11 from Herpes Simplex Virus 1 Is an Intrinsically Disordered, RNA-Binding Protein.
mBio 11(3) (2020)
Metrick CM, Koenigsberg AL, Heldwein EE
|
| RgGuinier |
2.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
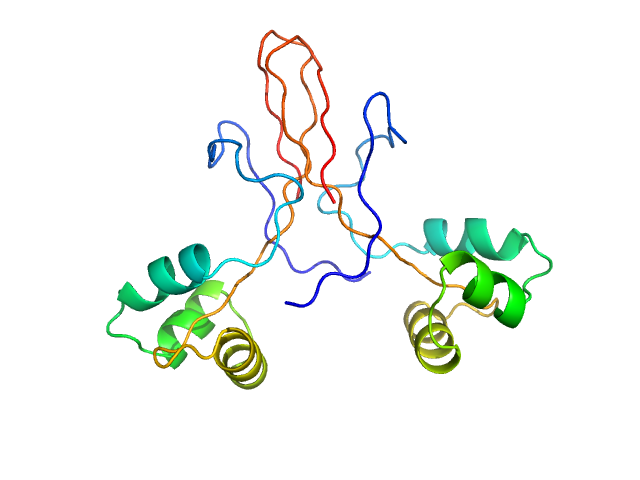
| Sample: |
Phosphoprotein tetramer, 317 kDa Nipah henipavirus protein
|
| Buffer: |
20 mM Tris-HCL, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2011 Nov 3
|
Structural Description of the Nipah Virus Phosphoprotein and Its Interaction with STAT1.
Biophys J (2020)
Jensen MR, Yabukarski F, Communie G, Condamine E, Mas C, Volchkova V, Tarbouriech N, Bourhis JM, Volchkov V, Blackledge M, Jamin M
|
| RgGuinier |
10.9 |
nm |
| Dmax |
39.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
Protein ninH dimer, 20 kDa Escherichia phage lambda protein
|
| Buffer: |
150 mM NaCl, 50 mM phosphate buffer (pH 7.4), 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 1
|
A bacteriophage mimic of the bacterial nucleoid-associated protein Fis.
Biochem J (2020)
Chakraborti S, Balakrishnan D, Trotter AJ, Gittens WH, Yang AWH, Jolma A, Paterson JR, Świątek S, Plewka J, Curtis FA, Bowers LY, Pålsson LO, Hughes TR, Taube M, Kozak M, Heddle JG, Sharples GJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2015 May 24
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2015 Sep 21
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|