|
|
|
|

|
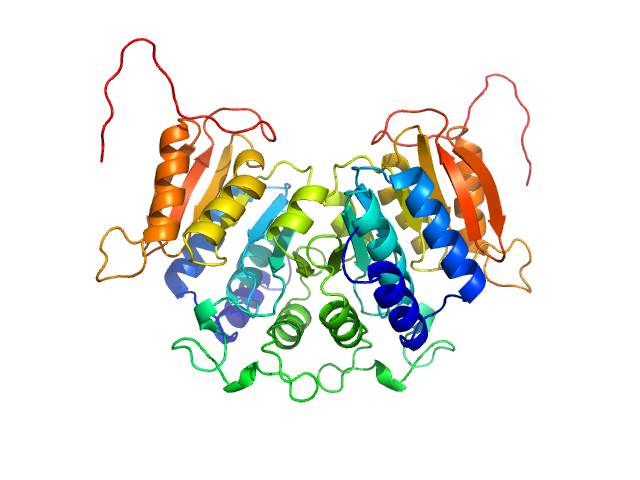
| Sample: |
Diadenylate cyclase dimer, 39 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
PLoS Pathog 15(1):e1007537 (2019)
Tosi T, Hoshiga F, Millership C, Singh R, Eldrid C, Patin D, Mengin-Lecreulx D, Thalassinos K, Freemont P, Gründling A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
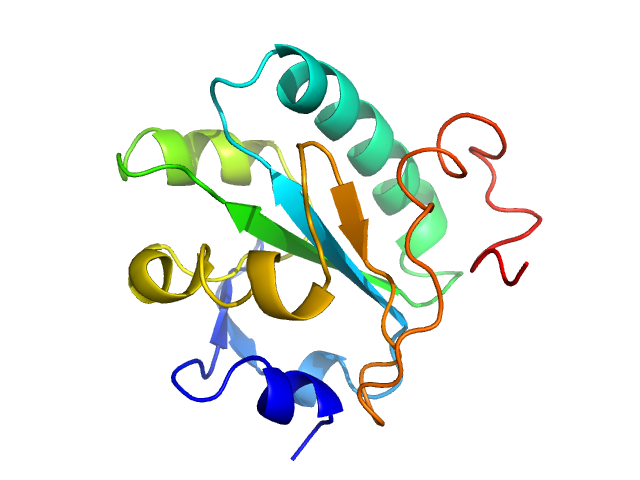
| Sample: |
Methyltransferase domain protein dimer, 51 kDa Mycolicibacterium hassiacum protein
|
| Buffer: |
20 mM bis-tris propane, 50 mM NaCl, 2 mM DTT, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jun 21
|
Biosynthesis of mycobacterial methylmannose polysaccharides requires a unique 1-O-methyltransferase specific for 3-O-methylated mannosides.
Proc Natl Acad Sci U S A 116(3):835-844 (2019)
Ripoll-Rozada J, Costa M, Manso JA, Maranha A, Miranda V, Sequeira A, Ventura MR, Macedo-Ribeiro S, Pereira PJB, Empadinhas N
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tryparedoxin monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|

|
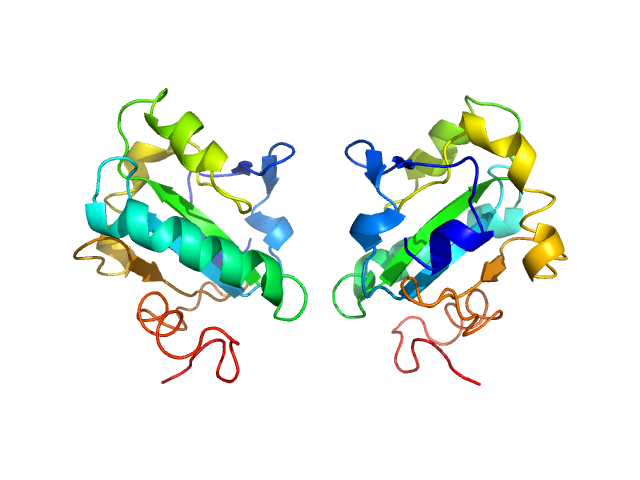
| Sample: |
Tryparedoxin monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
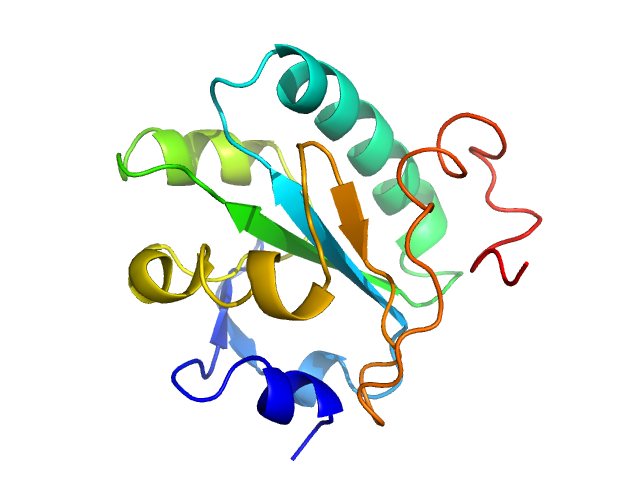
| Sample: |
Tryparedoxin, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tryparedoxin W39A monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
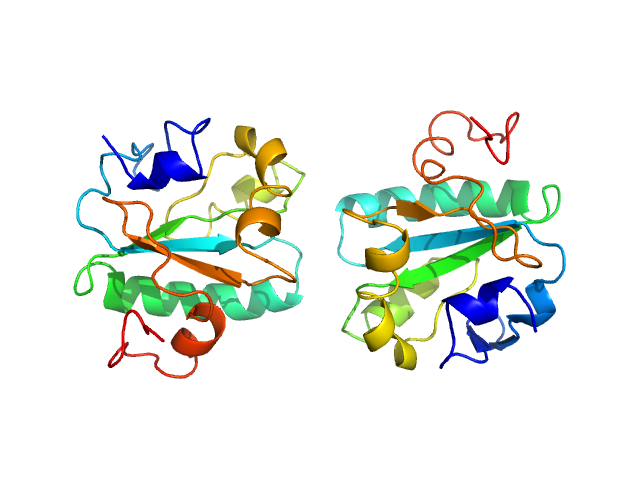
| Sample: |
Tryparedoxin W39A monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
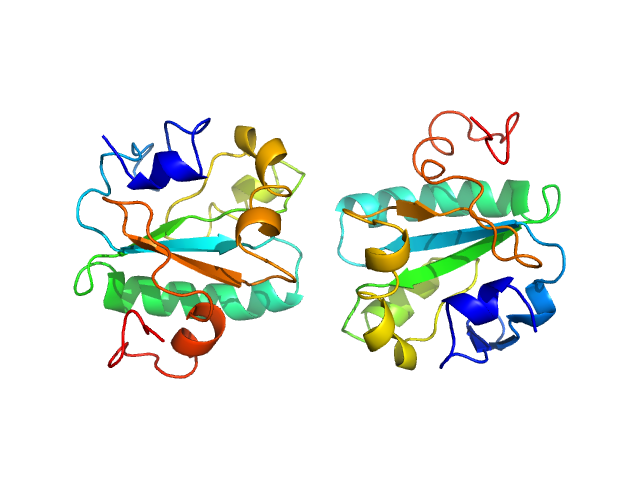
| Sample: |
Tryparedoxin W39A, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
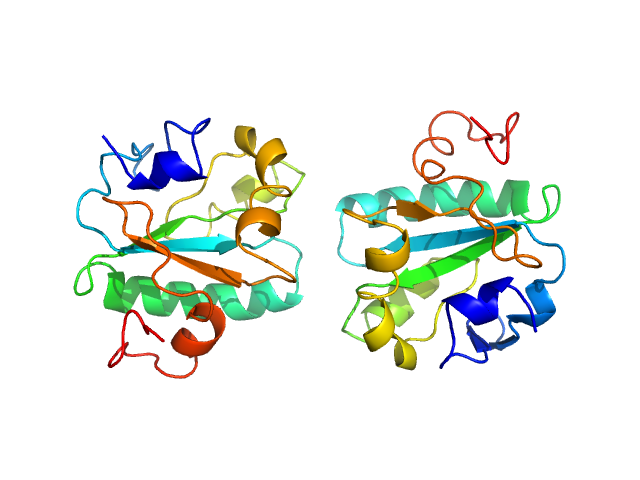
| Sample: |
Tryparedoxin W70A monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.9 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
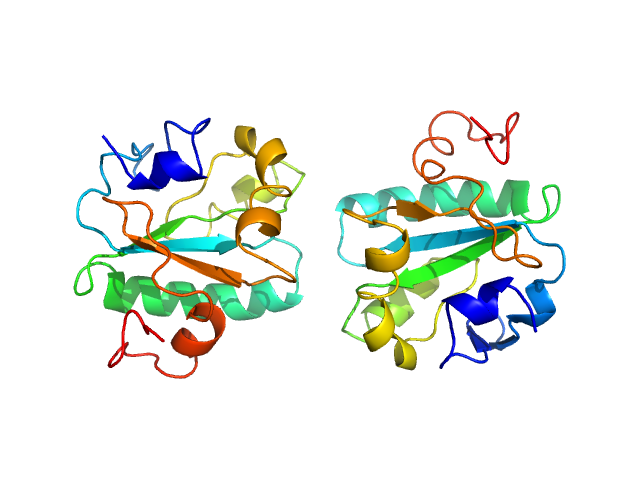
| Sample: |
Tryparedoxin W70A monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
31 |
nm3 |
|
|