|
|
|
|
|
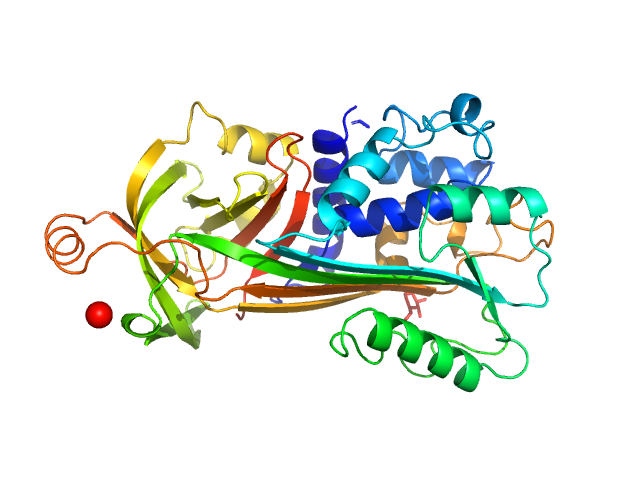
| Sample: |
Type II secretion system protein L, periplasmic domain dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
50 mM TRIS, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 8
|
Structure and oligomerization of the periplasmic domain of GspL from the type II secretion system of Pseudomonas aeruginosa.
Sci Rep 8(1):16760 (2018)
Fulara A, Vandenberghe I, Read RJ, Devreese B, Savvides SN
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Jan 24
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
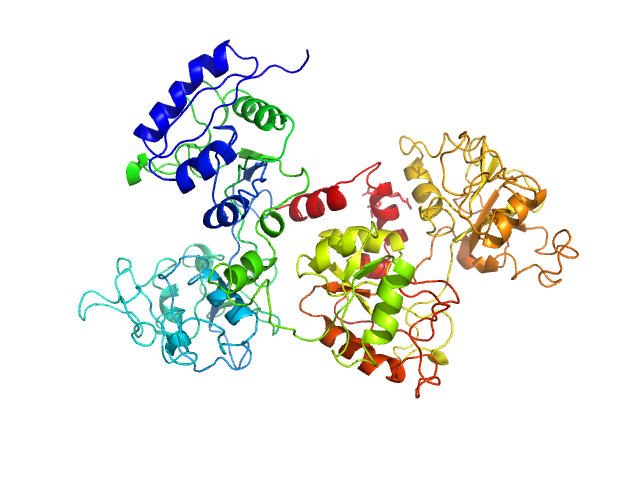
| Sample: |
Ovotransferrin monomer, 76 kDa Gallus gallus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Jan 24
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
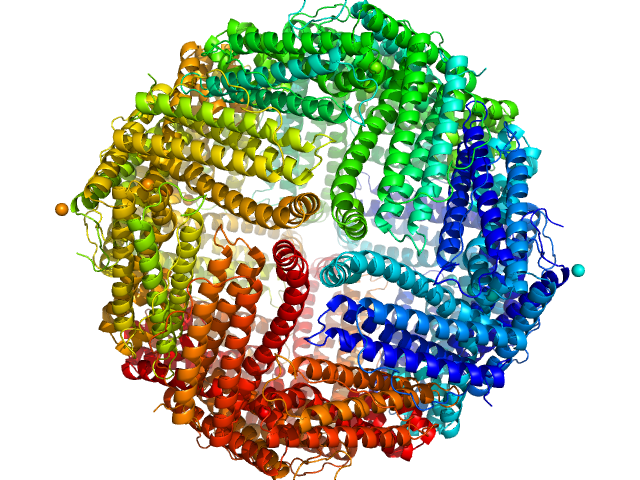
| Sample: |
Ferritin light chain 24-mer, 476 kDa Equus caballus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Jan 24
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
5.1 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
685 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Old Yellow Enzyme of Leishmania braziliensis monomer, 42 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris-HCl 100 mM NaCl and 1 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2015 May 26
|
Structural studies of Old Yellow Enzyme of Leishmania braziliensis in solution.
Arch Biochem Biophys (2018)
Walbert Veloso-Silva LL, Dores-Silva PR, Bertolino Reis DE, Moreno Oliveira LF, Libardi SH, Borges JC
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
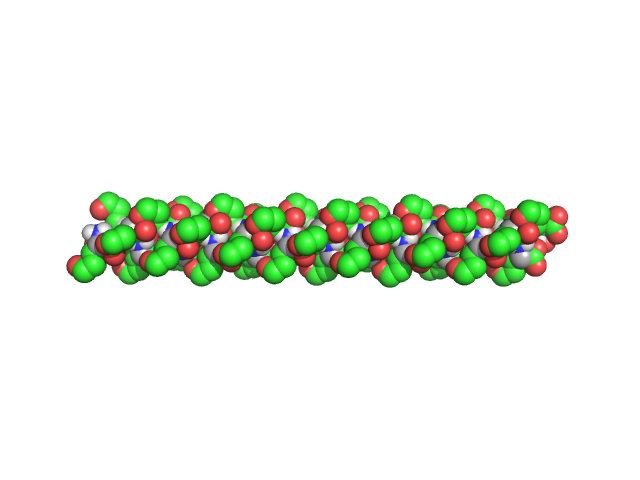
| Sample: |
Poly-L-Glutamic Acid, 6 kDa
|
| Buffer: |
Dimethyl Sulfoxide (DMSO), pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 11
|
β2-Type Amyloidlike Fibrils of Poly-l-glutamic Acid Convert into Long, Highly Ordered Helices upon Dissolution in Dimethyl Sulfoxide.
J Phys Chem B 122(50):11895-11905 (2018)
Berbeć S, Dec R, Molodenskiy D, Wielgus-Kutrowska B, Johannessen C, Hernik-Magoń A, Tobias F, Bzowska A, Ścibisz G, Keiderling TA, Svergun D, Dzwolak W
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
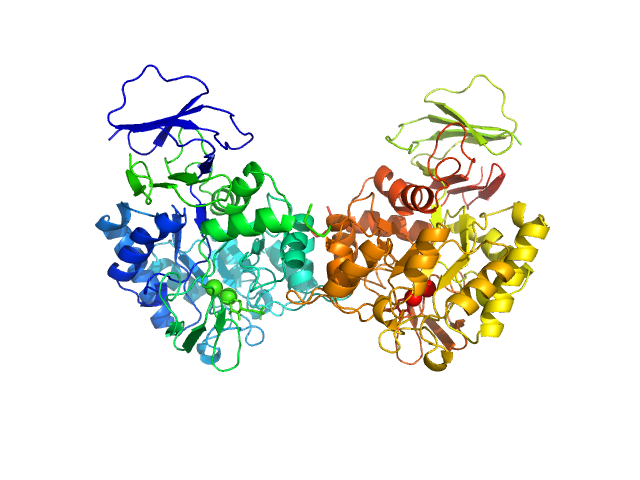
|
| Sample: |
N-acetylglucosamine-6-phosphate deacetylase dimer, 86 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 26
|
Functional and solution structure studies of amino sugar deacetylase and deaminase enzymes from Staphylococcus aureus.
FEBS Lett (2018)
Davies JS, Coombes D, Horne CR, Pearce FG, Friemann R, North RA, Dobson RCJ
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
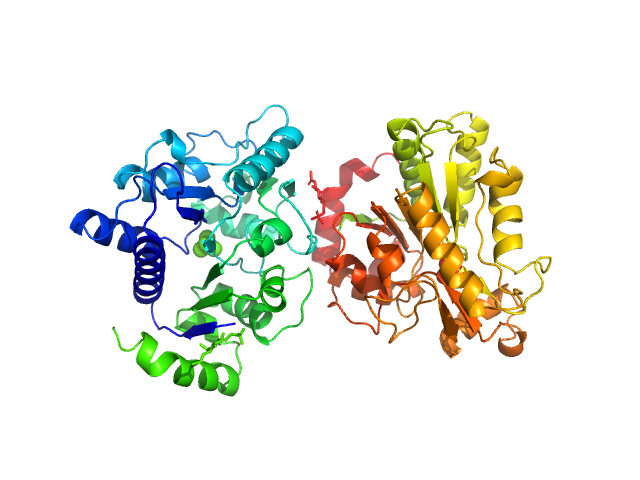
|
| Sample: |
Glucosamine-6-phosphate deaminase dimer, 57 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 26
|
Functional and solution structure studies of amino sugar deacetylase and deaminase enzymes from Staphylococcus aureus.
FEBS Lett (2018)
Davies JS, Coombes D, Horne CR, Pearce FG, Friemann R, North RA, Dobson RCJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
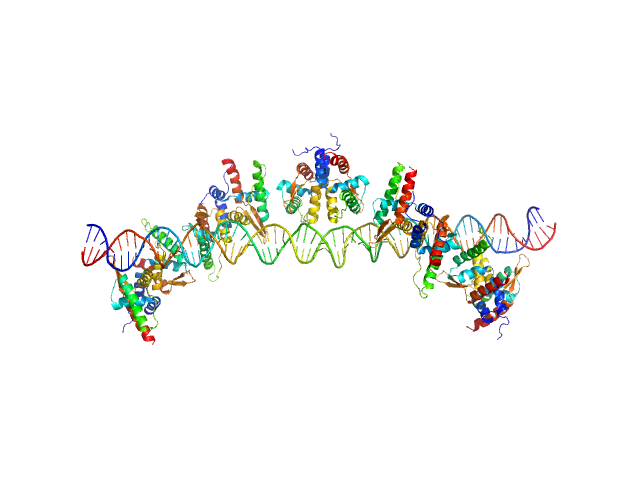
|
| Sample: |
S48 DNA strand 1 monomer, 21 kDa DNA
S48 DNA strand 2 monomer, 21 kDa DNA
TubR of the pXO1-like plasmid pBc10987 from B. cereus (Bc-TubR) decamer, 137 kDa protein
|
| Buffer: |
0.1 M NaCl, 10 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Nov 28
|
Cooperative DNA Binding of the Plasmid Partitioning Protein TubR from the Bacillus cereus pXO1 Plasmid.
J Mol Biol (2018)
Hayashi I, Oda T, Sato M, Fuchigami S
|
| RgGuinier |
6.1 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
305 |
nm3 |
|
|
|
|
|
|
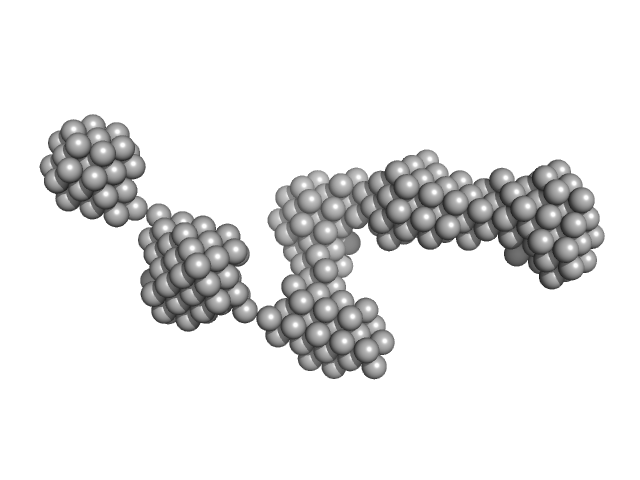
|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
19 |
nm3 |
|
|