|
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Ovis aries protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 4
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Ovis aries protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, 0.001 mM CuSO4, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 4
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Ovis aries protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 4
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Ovis aries protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, 0.001 mM CuSO4, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 4
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
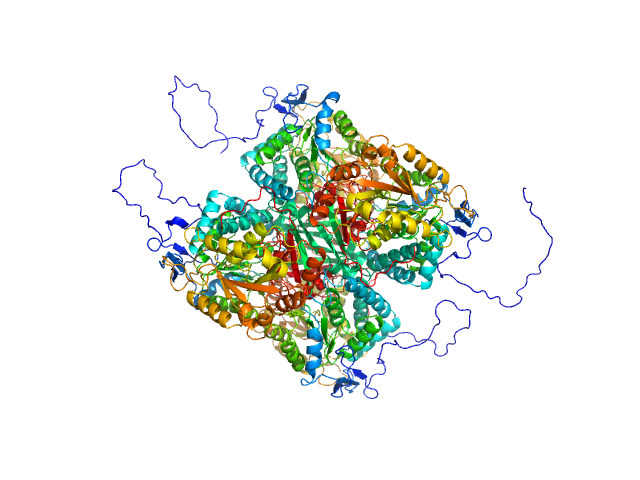
| Sample: |
Aldehyde dehydrogenase 12 tetramer, 242 kDa Zea mays protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM TCEP, and 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 6
|
Structural and Biochemical Characterization of Aldehyde Dehydrogenase 12, the Last Enzyme of Proline Catabolism in Plants.
J Mol Biol (2018)
Korasick DA, Končitíková R, Kopečná M, Hájková E, Vigouroux A, Moréra S, Becker DF, Šebela M, Tanner JJ, Kopečný D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
351 |
nm3 |
|
|
|
|
|
|
|
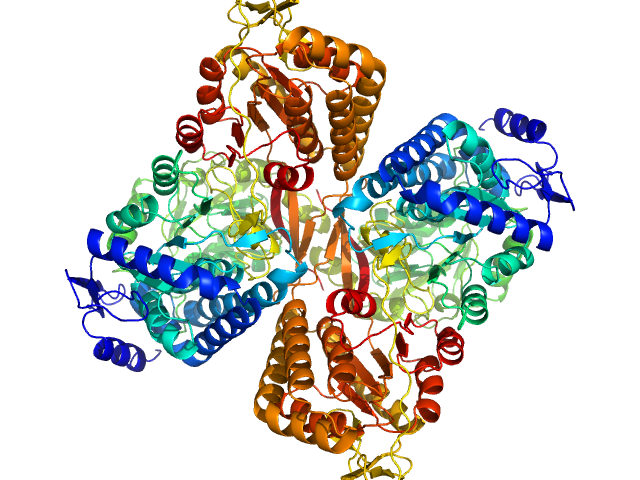
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
|
|
|
|
|
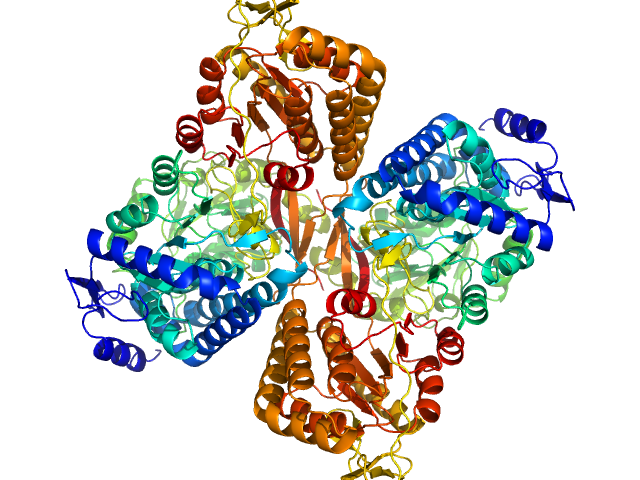
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|
|
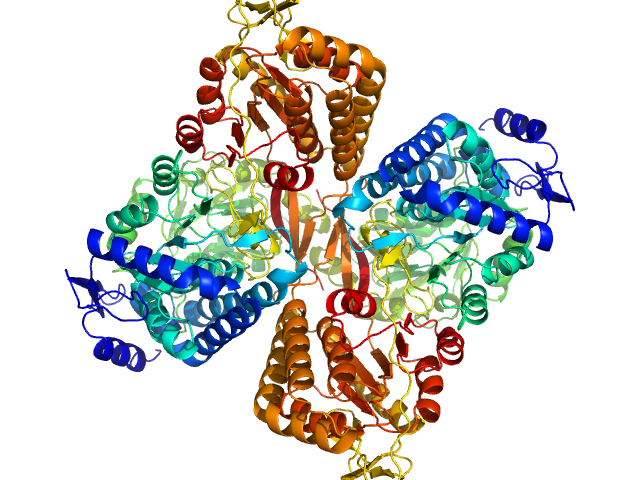
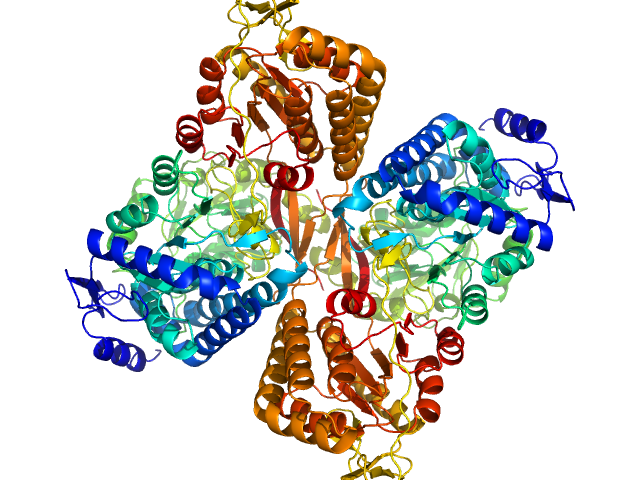
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
205 |
nm3 |
|
|
|
|
|
|
|
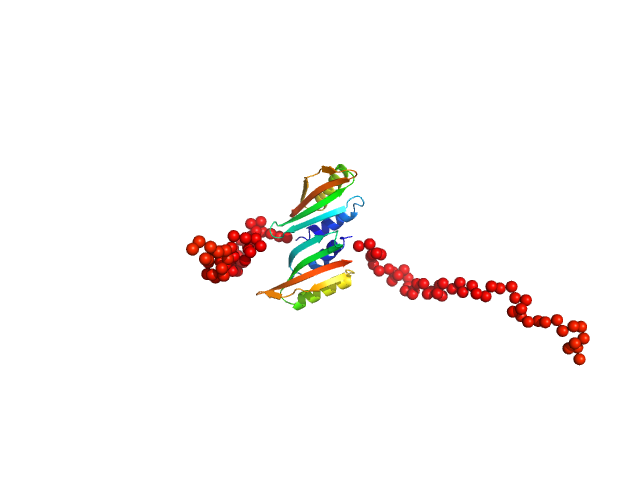
| Sample: |
Type II secretion system protein L, periplasmic domain dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
50 mM TRIS, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 8
|
Structure and oligomerization of the periplasmic domain of GspL from the type II secretion system of Pseudomonas aeruginosa.
Sci Rep 8(1):16760 (2018)
Fulara A, Vandenberghe I, Read RJ, Devreese B, Savvides SN
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
|
|