|
|
|
|
|
| Sample: |
E13A monomer, 17 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E13B monomer, 10 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E13C monomer, 6 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.3 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
|
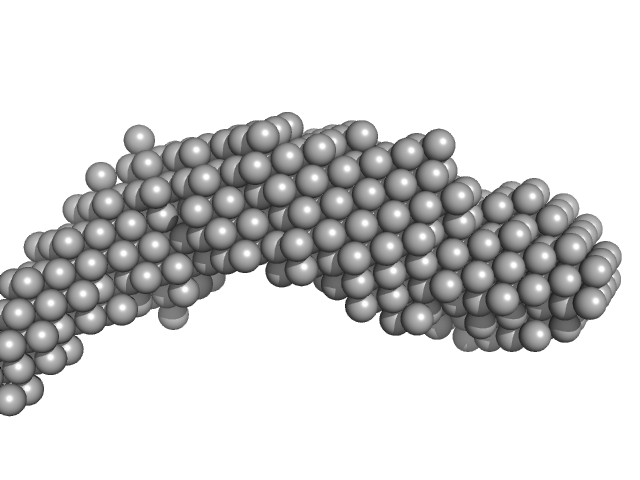
| Sample: |
E13AB monomer, 79 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
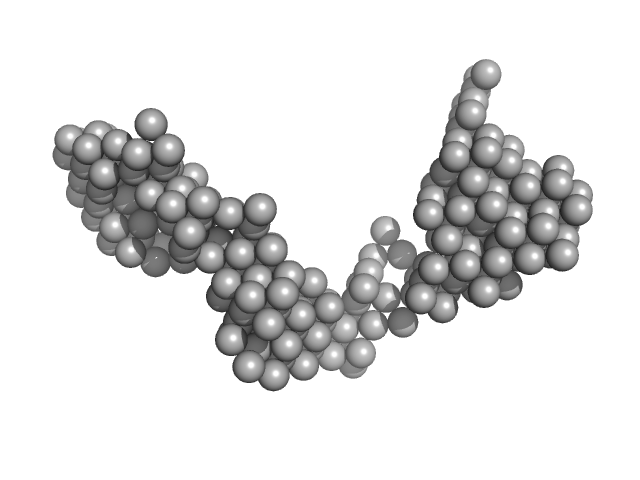
| Sample: |
E13ABC monomer, 42 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|

|
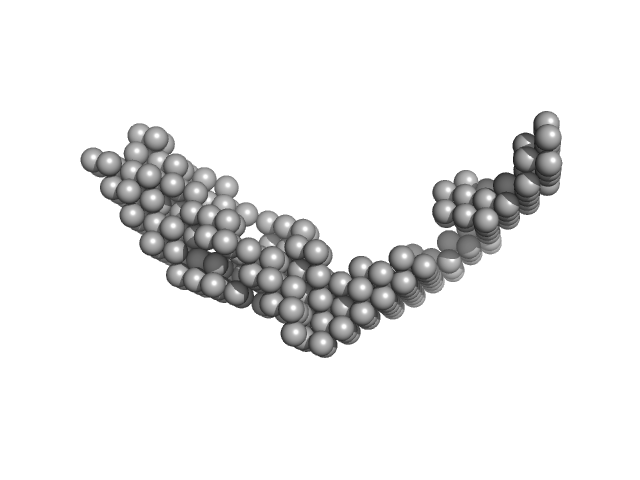
| Sample: |
E13Ae14Be13C monomer, 40 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E14Ae13Be14C monomer, 31 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nonstructural protein sigma NS octamer, 325 kDa Avian orthoreovirus protein
20mer RNA (unstructured) dimer, 13 kDa RNA
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 25
|
Stability of local secondary structure determines selectivity of viral RNA chaperones.
Nucleic Acids Res (2018)
Bravo JPK, Borodavka A, Barth A, Calabrese AN, Mojzes P, Cockburn JJB, Lamb DC, Tuma R
|
| RgGuinier |
7.8 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
964 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nonstructural protein sigma NS hexamer, 244 kDa Avian orthoreovirus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 25
|
Stability of local secondary structure determines selectivity of viral RNA chaperones.
Nucleic Acids Res (2018)
Bravo JPK, Borodavka A, Barth A, Calabrese AN, Mojzes P, Cockburn JJB, Lamb DC, Tuma R
|
| RgGuinier |
5.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
670 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Extender PN-Block (HL4) monomer, 27 kDa de novo protein protein
Stopper PN-Block (WA20) dimer, 25 kDa de novo protein protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 200 mM ArgHCl, 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2014 Dec 19
|
Self-Assembling Supramolecular Nanostructures Constructed from de Novo Extender Protein Nanobuilding Blocks.
ACS Synth Biol 7(5):1381-1394 (2018)
Kobayashi N, Inano K, Sasahara K, Sato T, Miyazawa K, Fukuma T, Hecht MH, Song C, Murata K, Arai R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
|
|