|
|
|
|
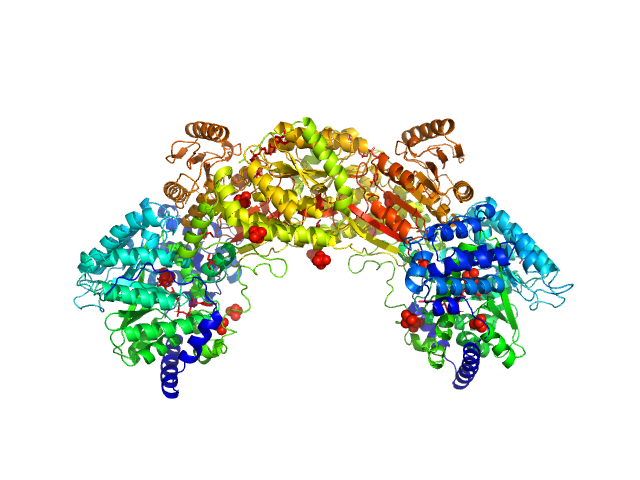
|
| Sample: |
Bifunctional protein PutA dimer, 215 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM TCEP, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 16
|
Redox Modulation of Oligomeric State in Proline Utilization A.
Biophys J 114(12):2833-2843 (2018)
Korasick DA, Campbell AC, Christgen SL, Chakravarthy S, White TA, Becker DF, Tanner JJ
|
| RgGuinier |
4.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
324 |
nm3 |
|
|
|
|
|
|
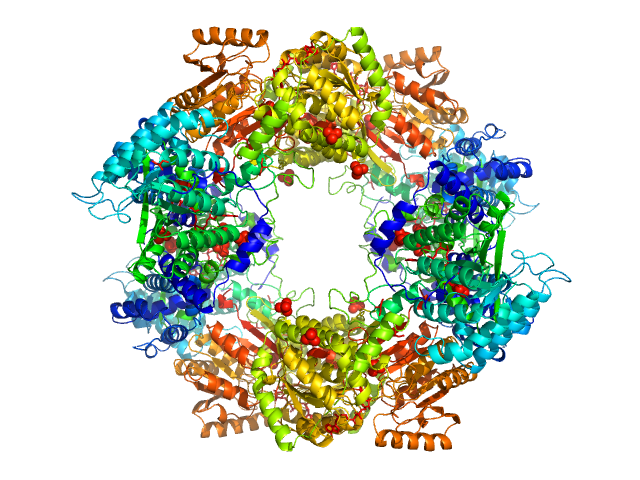
|
| Sample: |
Bifunctional protein PutA tetramer, 430 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM TCEP, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 16
|
Redox Modulation of Oligomeric State in Proline Utilization A.
Biophys J 114(12):2833-2843 (2018)
Korasick DA, Campbell AC, Christgen SL, Chakravarthy S, White TA, Becker DF, Tanner JJ
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
582 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lipase B from Pseudozyma antarctica, 33 kDa Moesziomyces antarcticus protein
|
| Buffer: |
100 mM NaCl, 20 mM Na2HPO4, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
|
|
|
|
|
|
|
| Sample: |
Lipase B from Pseudozyma antarctica, 33 kDa Moesziomyces antarcticus protein
|
| Buffer: |
100 mM NaCl, 20 mM Na2HPO4, 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
|
|
|
|
|
|
|
| Sample: |
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
E14A monomer, 21 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
E14B monomer, 13 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.9 |
nm |
| Dmax |
9.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
E14C monomer, 5 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.3 |
nm |
| Dmax |
5.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
E14AB monomer, 93 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
5.4 |
nm |
| Dmax |
24.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
E14ABC monomer, 106 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
4.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
152 |
nm3 |
|
|