|
|
|
|

|
| Sample: |
KLBS1-2 DNA monomer, 24 kDa unidentified herpesvirus DNA
ORF73 tetramer tetramer, 63 kDa Human herpesvirus 8 protein
ORF73 octamer octamer, 126 kDa Human herpesvirus 8 protein
KLBS1-2 DNA two monomers dimer, 48 kDa unidentified herpesvirus RNA
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Varkud Satellite (VS) ribozyme dimer, 120 kDa Neurospora RNA
|
| Buffer: |
10 mM Tris 25 mM KCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Mar 6
|
Crystal structure of the Varkud satellite ribozyme.
Nat Chem Biol 11(11):840-6 (2015)
Suslov NB, DasGupta S, Huang H, Fuller JR, Lilley DM, Rice PA, Piccirilli JA
|
| RgGuinier |
3.9 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
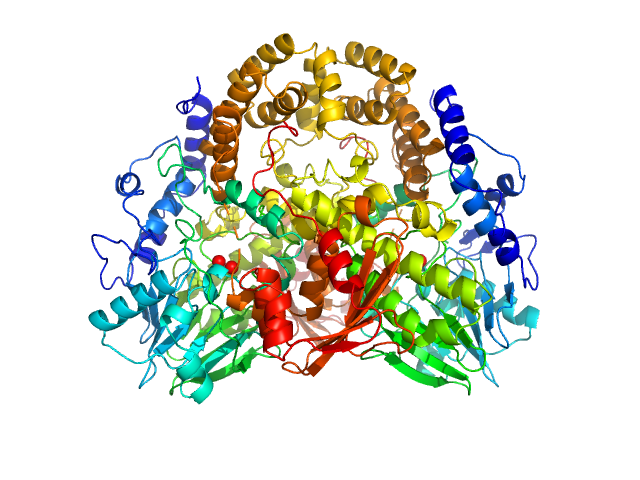
|
| Sample: |
Pseudomonas aeruginosa SDS hydrolase SdsA1 dimer, 145 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
50 mM HEPES, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2013 Jul 22
|
SdsA polymorph isolation and improvement of their crystal quality using nonconventional crystallization techniques
Journal of Applied Crystallography 48(5):1551-1559 (2015)
De la Mora E, Flores-Hernández E, Jakoncic J, Stojanoff V, Siliqi D, Sánchez-Puig N, Moreno A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
261 |
nm3 |
|
|
|
|
|
|
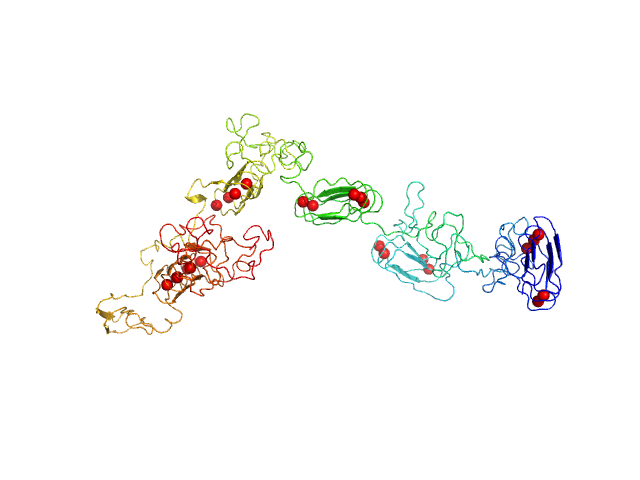
|
| Sample: |
RD domain of B. Pertussis Adenylate Cyclase Toxin (CyaA) monomer, 73 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM DTT, 4 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 May 31
|
Structural models of intrinsically disordered and calcium-bound folded states of a protein adapted for secretion.
Sci Rep 5:14223 (2015)
O'Brien DP, Hernandez B, Durand D, Hourdel V, Sotomayor-Pérez AC, Vachette P, Ghomi M, Chamot-Rooke J, Ladant D, Brier S, Chenal A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
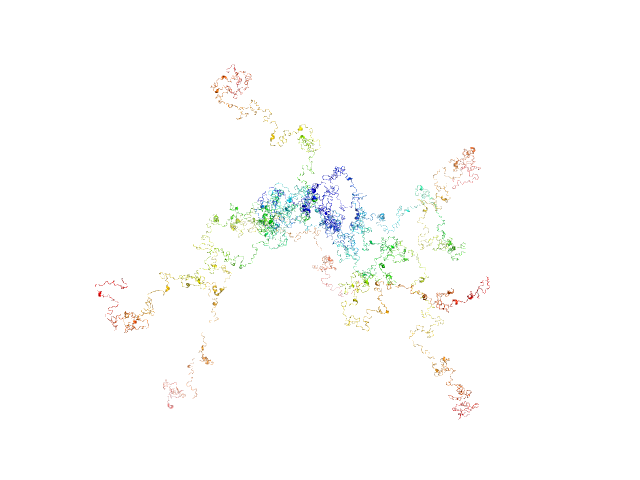
|
| Sample: |
RD domain of B. Pertussis Adenylate Cyclase Toxin (CyaA) monomer, 73 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 May 31
|
Structural models of intrinsically disordered and calcium-bound folded states of a protein adapted for secretion.
Sci Rep 5:14223 (2015)
O'Brien DP, Hernandez B, Durand D, Hourdel V, Sotomayor-Pérez AC, Vachette P, Ghomi M, Chamot-Rooke J, Ladant D, Brier S, Chenal A
|
| RgGuinier |
8.3 |
nm |
| Dmax |
33.0 |
nm |
|
|
|
|
|
|
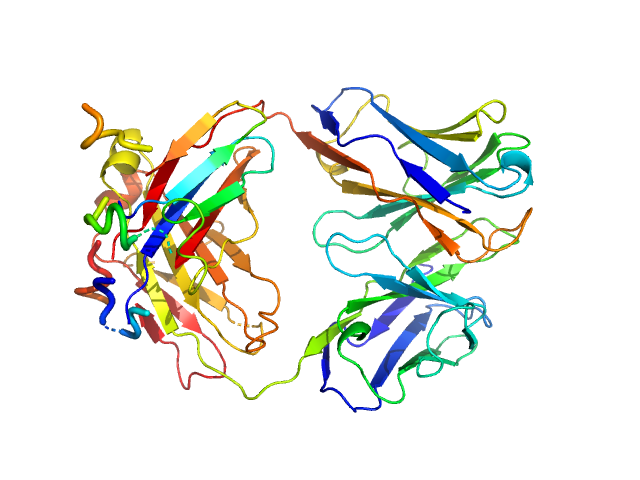
|
| Sample: |
Anti-TG2 antibody (679 14 E06) monomer, 48 kDa protein
|
| Buffer: |
20 mM Tris 150mM NaCl 1mM EDTA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jan 17
|
Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J Biol Chem 290(35):21365-75 (2015)
Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Antiapoptotic membrane protein dimer, 39 kDa Deerpox virus W-1170-84 protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Structural basis of Deerpox virus-mediated inhibition of apoptosis.
Acta Crystallogr D Biol Crystallogr 71(Pt 8):1593-603 (2015)
Burton DR, Caria S, Marshall B, Barry M, Kvansakul M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 20
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
|
|
|
|
|
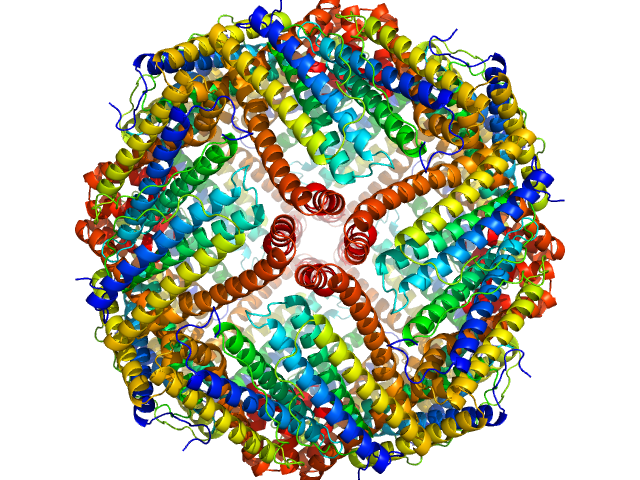
| Sample: |
Ferritin light chain 24-mer, 479 kDa Equus caballus protein
|
| Buffer: |
PBS, 50% Glycerol, 0.076 M NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 20
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
6.8 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
619 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ovotransferrin monomer, 78 kDa Gallus gallus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 21
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|