|
|
|
|
|
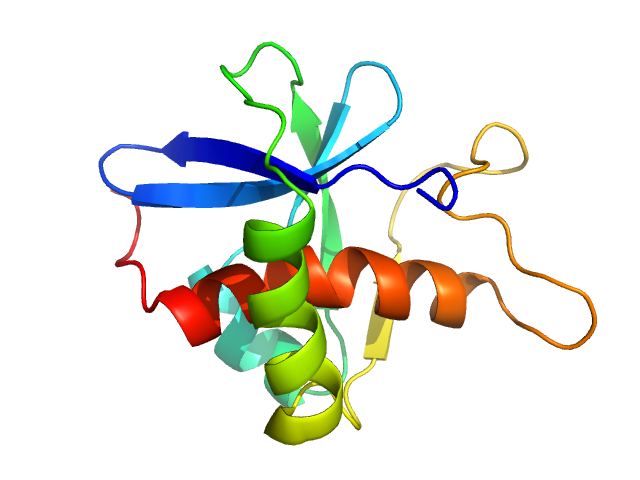
| Sample: |
CrTPpart2 of cysteine synthase A (CysK) monomer, 16 kDa Paulinella chromatophora protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
The Paulinella
chromatophore transit peptide part2 adopts a structural fold similar to the γ-glutamyl-cyclotransferase fold
Plant Physiology (2025)
Klimenko V, Reiners J, Applegate V, Reimann K, Popowicz G, Hoeppner A, Papadopoulos A, Smits S, Nowack E
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
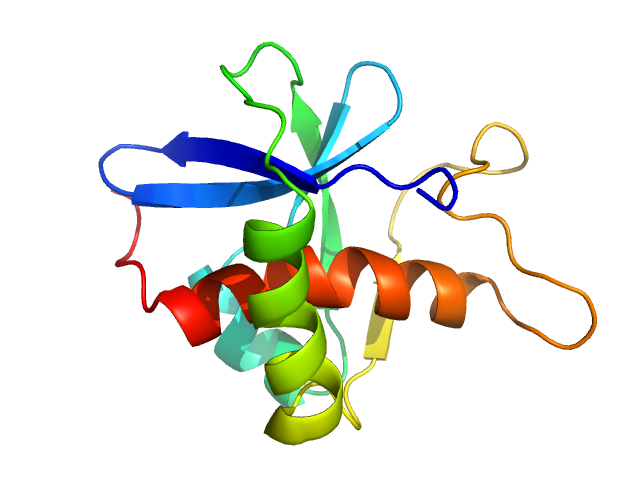
| Sample: |
Genome polyprotein monomer, 13 kDa Turkey avisivirus protein
|
| Buffer: |
20 mM citrate pH 5.4, 150 mM NaCl, 2 mM TCEP, 1% glycerol, pH: 5.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 2
|
Solution structure and functional characterization of an avian picornavirus 2AH/NC protein
Anna Wehlin
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
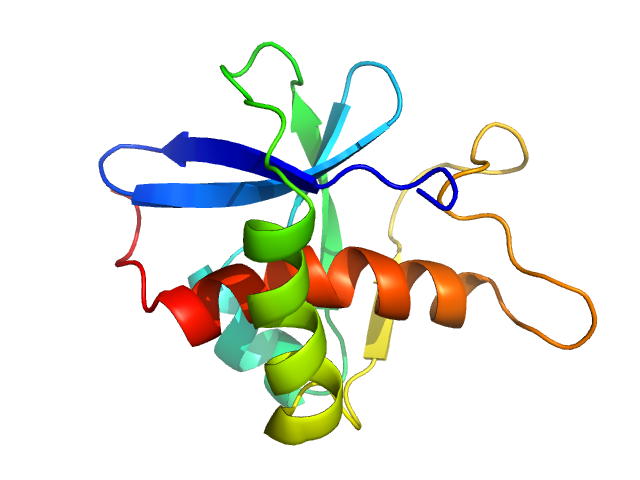
| Sample: |
Genome polyprotein monomer, 13 kDa Turkey avisivirus protein
|
| Buffer: |
20mM Citrate buffer pH 6.5, 150mM NaCl, 2mM TCEP, 1% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 May 15
|
Solution structure and functional characterization of an avian picornavirus 2AH/NC protein
Anna Wehlin
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
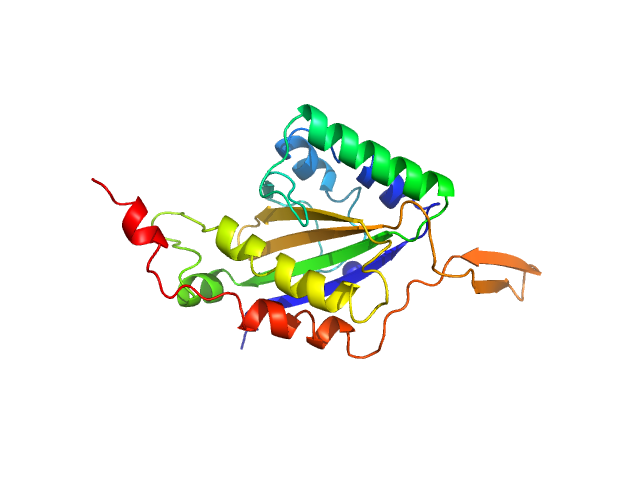
| Sample: |
Genome polyprotein monomer, 13 kDa Turkey avisivirus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TECP, 1% Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 2
|
Solution structure and functional characterization of an avian picornavirus 2AH/NC protein
Anna Wehlin
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
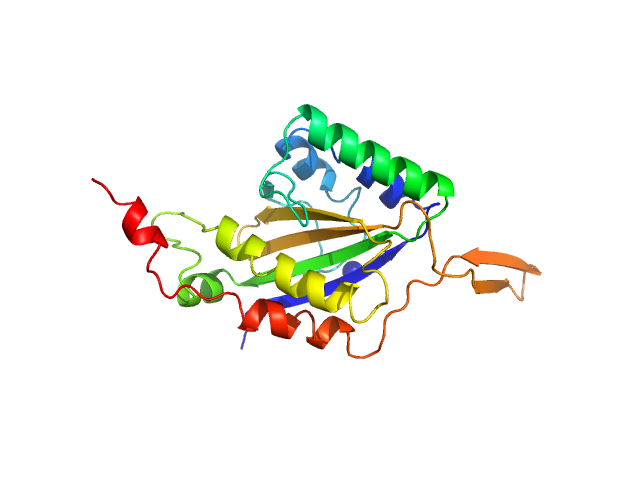
| Sample: |
Relaxase monomer, 29 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
Structure of relaxase from Bacillus subtilis conjugative plasmid pLS20 reveals a new mechanism of DNA processing mediated by dimerization
Isidro Crespo
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Relaxase dimer, 57 kDa Bacillus subtilis subsp. … protein
Nic sequence, single stranded dimer, 12 kDa Bacillus subtilis DNA
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
Structure of relaxase from Bacillus subtilis conjugative plasmid pLS20 reveals a new mechanism of DNA processing mediated by dimerization
Isidro Crespo
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
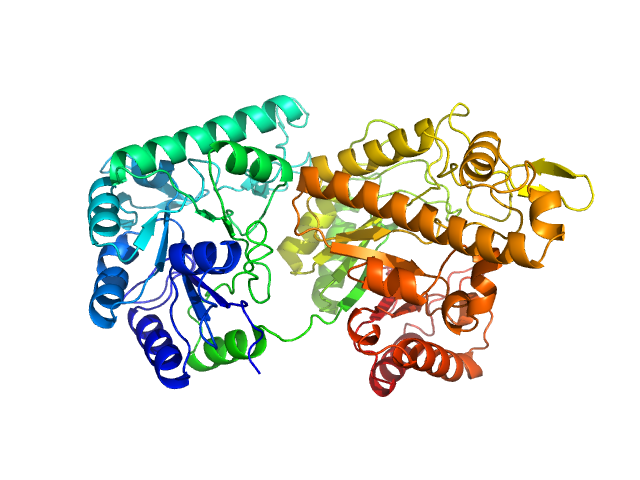
| Sample: |
Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase PgaB monomer, 73 kDa Serratia marcescens protein
|
| Buffer: |
phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Feb 3
|
Structural, biophysical and biochemical studies of CAZYmes involved in exopolysaccharides degradation of microbial biofilms
Universidade de São Paulo Master's thesis (2025)
Amanda Freitas Cruz
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase PgaB monomer, 69 kDa Serratia marcescens protein
|
| Buffer: |
phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Feb 3
|
Structural, biophysical and biochemical studies of CAZYmes involved in exopolysaccharides degradation of microbial biofilms
Universidade de São Paulo Master's thesis (2025)
Amanda Freitas Cruz
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ABC transporter, permease protein, extracellular domain monomer, 23 kDa Streptococcus agalactiae serotype … protein
|
| Buffer: |
25 mM MES, 500 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 3
|
The extracellular domain of SaNSrFP binds bacitracin and allows the identification of new members of the BceAB transporter family
Frontiers in Microbiology 16 (2025)
Mammen C, Gottstein J, Cea P, Tantsur K, Reiners J, Bonus M, Gohlke H, Smits S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ABC transporter ATP-binding protein dimer, 62 kDa Streptococcus agalactiae protein
ABC transporter, permease protein monomer, 74 kDa Streptococcus agalactiae serotype … protein
|
| Buffer: |
50 mM Tris, 300 mM NaCl, 10 % Glycerol, 0.005 % LMNG,, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Apr 25
|
The extracellular domain of SaNSrFP binds bacitracin and allows the identification of new members of the BceAB transporter family
Frontiers in Microbiology 16 (2025)
Mammen C, Gottstein J, Cea P, Tantsur K, Reiners J, Bonus M, Gohlke H, Smits S
|
| RgGuinier |
5.8 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
735 |
nm3 |
|
|