|
|
|
|
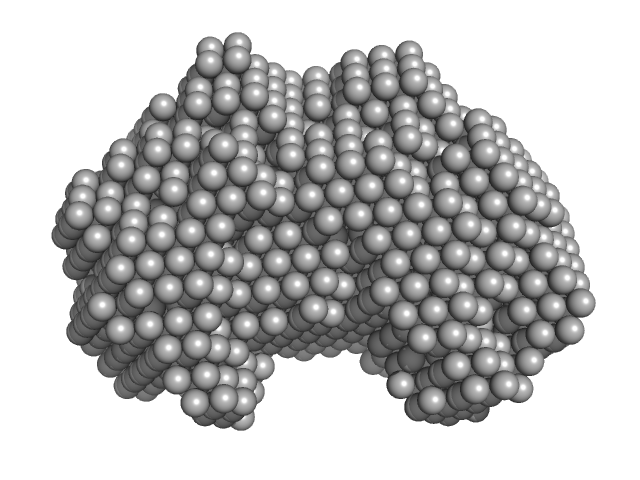
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 9
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 9
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 19
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
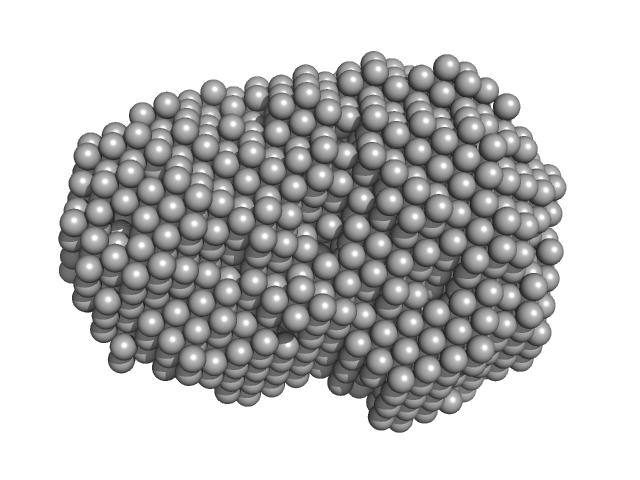
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
IMP-1 saRNA monomer, 3720 kDa RNA
|
| Buffer: |
5 mM Sodium Citrate, pH: 6.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 27
|
Biophysical characterisation of the structure of a SARS-CoV-2 self-amplifying—RNA (saRNA) vaccine
Biology Methods and Protocols (2023)
Myatt D, Wharram L, Graham C, Liddell J, Branton H, Pizzey C, Cowieson N, Rambo R, Shattock R
|
| RgGuinier |
23.8 |
nm |
| Dmax |
85.9 |
nm |
| VolumePorod |
6265 |
nm3 |
|
|
|
|
|
|
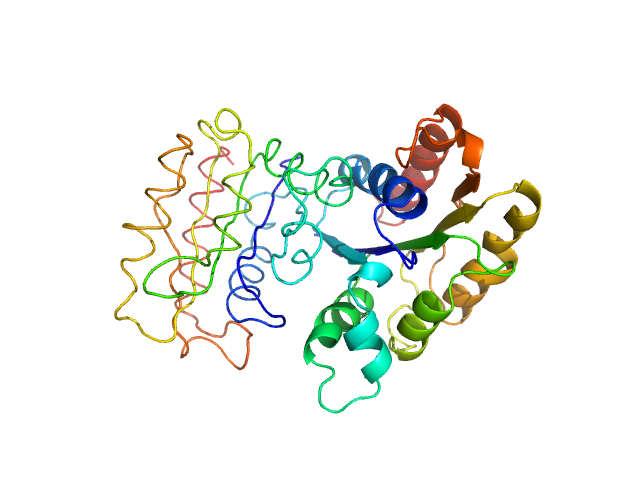
|
| Sample: |
NADPH-dependent FMN reductase dimer, 41 kDa Paracoccus denitrificans protein
|
| Buffer: |
50 mM sodium phosphate buffer, 300 mM NaCl, 500 mM imidazole, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 Feb 22
|
Structural Insight into Catalysis by the Flavin-Dependent NADH Oxidase (Pden_5119) of Paracoccus denitrificans.
Int J Mol Sci 24(4) (2023)
Kryl M, Sedláček V, Kučera I
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NURS complex subunit red1 monomer, 8 kDa Schizosaccharomyces pombe (strain … protein
|
| Buffer: |
20 mM HEPES, pH 7.5, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 12
|
Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun 14(1):772 (2023)
Soni K, Sivadas A, Horvath A, Dobrev N, Hayashi R, Kiss L, Simon B, Wild K, Sinning I, Fischer T
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
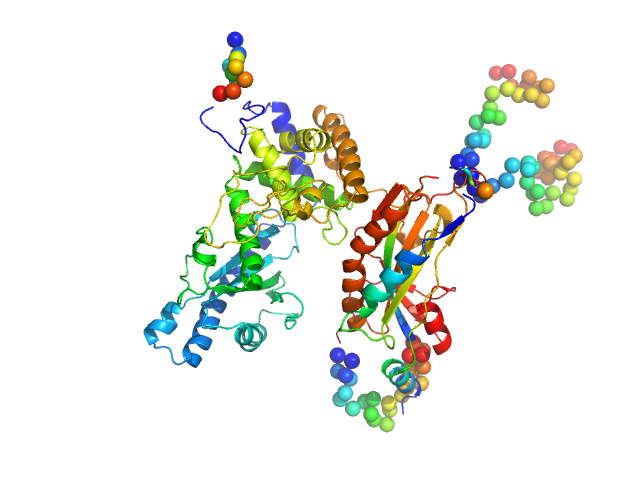
|
| Sample: |
NURS complex subunit red1 monomer, 8 kDa Schizosaccharomyces pombe (strain … protein
Poly(A) polymerase pla1 monomer, 65 kDa Schizosaccharomyces pombe (strain … protein
|
| Buffer: |
20 mM HEPES, pH 7.5, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 12
|
Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun 14(1):772 (2023)
Soni K, Sivadas A, Horvath A, Dobrev N, Hayashi R, Kiss L, Simon B, Wild K, Sinning I, Fischer T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
138 |
nm3 |
|
|