|
|
|
|
|
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp8 ammonium salt monomer, 2 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
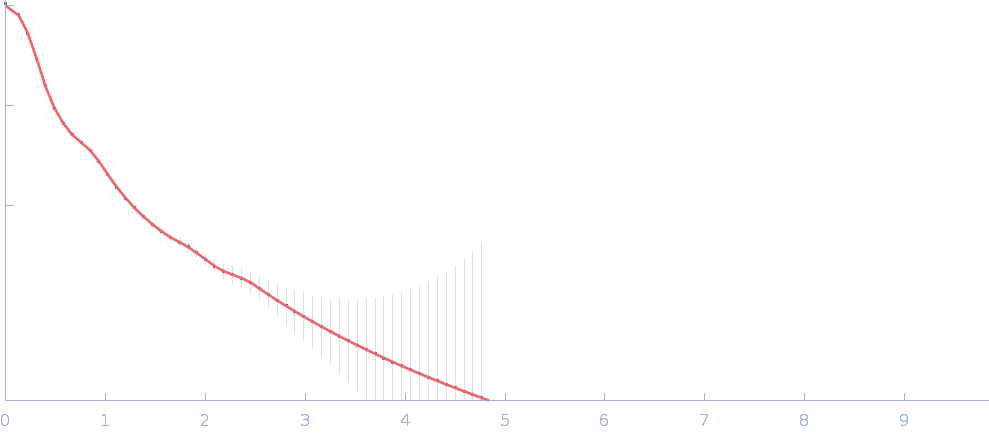
| RgGuinier |
6.5 |
nm |
| Dmax |
23.5 |
nm |
| VolumePorod |
729 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp8 ammonium salt monomer, 2 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
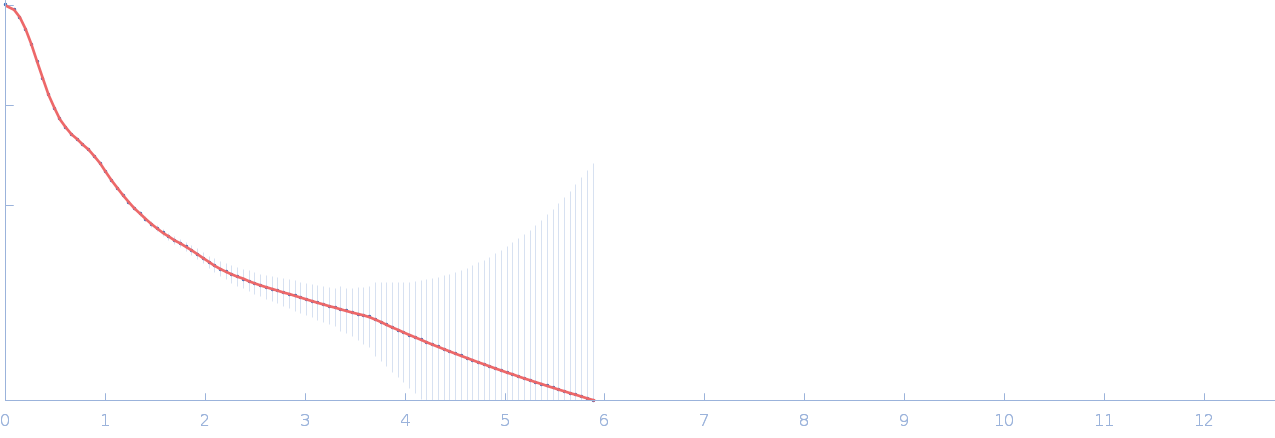
| RgGuinier |
6.4 |
nm |
| Dmax |
21.9 |
nm |
| VolumePorod |
723 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp10 ammonium salt monomer, 3 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
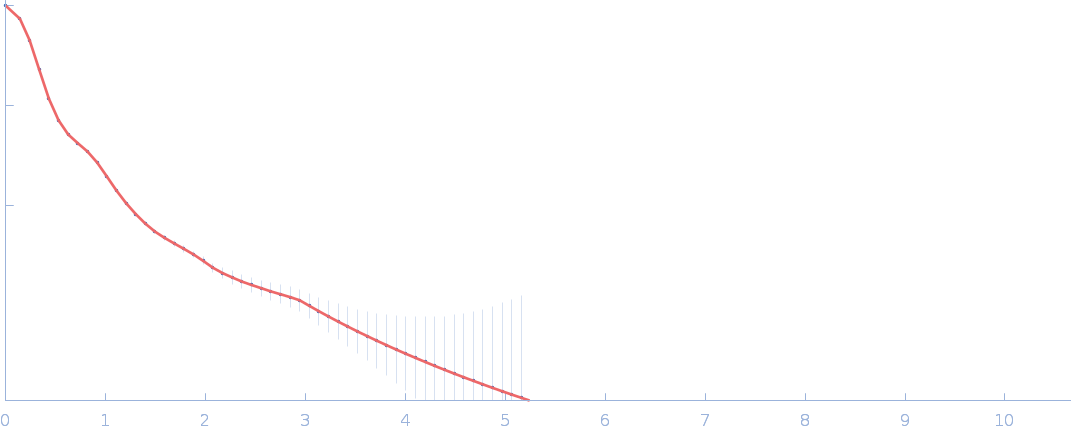
| RgGuinier |
6.4 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
758 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp10 ammonium salt monomer, 3 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
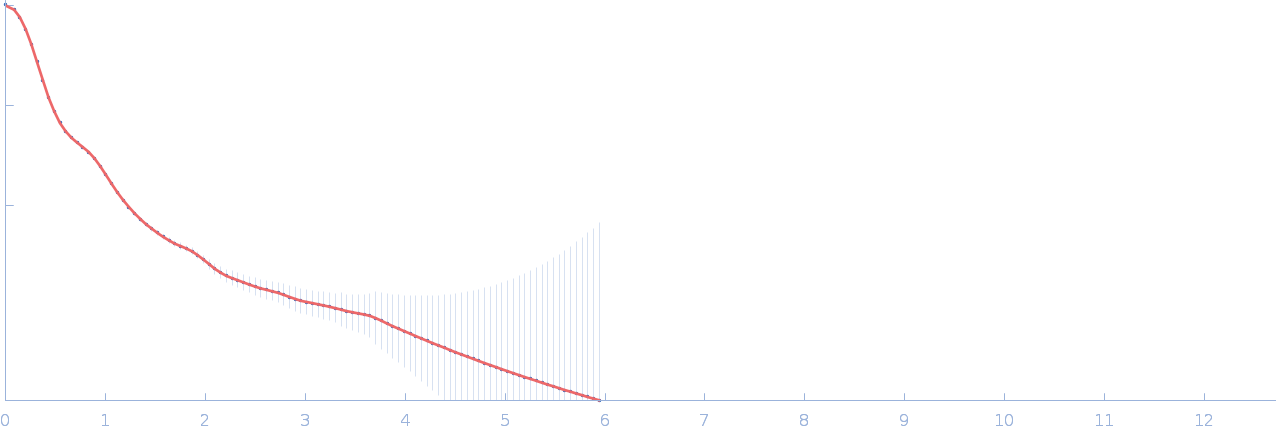
| RgGuinier |
6.5 |
nm |
| Dmax |
21.9 |
nm |
| VolumePorod |
769 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein TOC75, chloroplastic monomer, 32 kDa Pisum sativum protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|

|
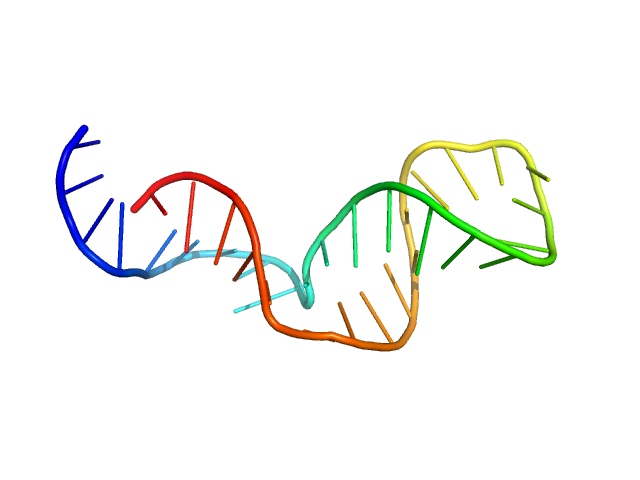
| Sample: |
AT-rich dsDNA monomer, 7 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome.
Life Sci Alliance 6(3) (2023)
Watson JA, Pantier R, Jayachandran U, Chhatbar K, Alexander-Howden B, Kruusvee V, Prendecki M, Bird A, Cook AG
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
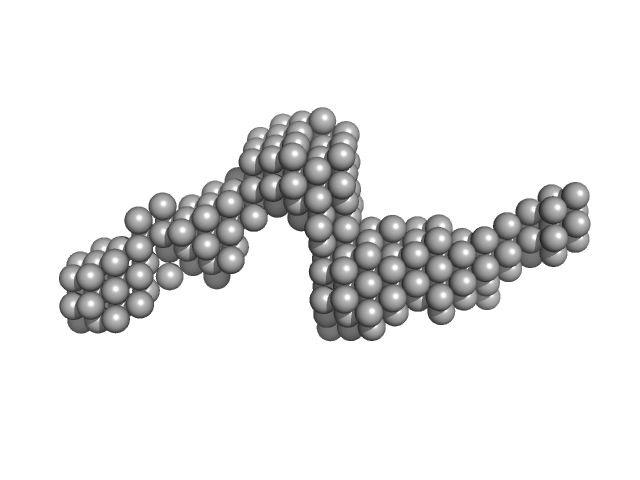
| Sample: |
Gli-233 - ssDNA aptamer to glioma brain tumor cells monomer, 10 kDa Artificially synthesized DNA
|
| Buffer: |
Dulbecco phosphate-buffered saline (DPBS, with Ca and Mg), pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 17
|
Development of DNA Aptamers for Visualization of Glial Brain Tumors and Detection of Circulating Tumor Cells
Molecular Therapy - Nucleic Acids (2023)
Kichkailo A, Narodov A, Komarova M, Zamay T, Zamay G, Kolovskaya O, Erakhtin E, Glazyrin Y, Veprintsev D, Moryachkov R, Zabluda V, Shchugoreva I, Artyushenko P, Mironov V, Morozov D, Khorzhevskii V, Gorbushin A, Koshmanova A, Nikolaeva E, Grinev I, Voronkovskii I, Grek D, Belugin K, Volzhentsev A, Badmaev O, Luzan N, Lukyanenko K, Peters G, Lapin I, Kirichenko A, Konarev P, Morozov E, Mironov G, Gargaun A, Muharemagic D, Zamay S, Kochkina E, Dymova M, Smolyarova T, Sokolov A, Modestov A, Tokarev N, Shepelevich N, Ozerskaya A, Chanchikova N, Krat A, Zukov R, Bakhtina V, Shnyakin P, Shesternya P, Svetlichnyi V, Petrova M, Artyukhov I, Tomilin F, Berezovski M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gli-55 - ssDNA aptamer specific to glioma brain tumor cells monomer, 19 kDa Artificially synthesized DNA
|
| Buffer: |
Dulbecco phosphate-buffered saline (DPBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 30
|
Development of DNA Aptamers for Visualization of Glial Brain Tumors and Detection of Circulating Tumor Cells
Molecular Therapy - Nucleic Acids (2023)
Kichkailo A, Narodov A, Komarova M, Zamay T, Zamay G, Kolovskaya O, Erakhtin E, Glazyrin Y, Veprintsev D, Moryachkov R, Zabluda V, Shchugoreva I, Artyushenko P, Mironov V, Morozov D, Khorzhevskii V, Gorbushin A, Koshmanova A, Nikolaeva E, Grinev I, Voronkovskii I, Grek D, Belugin K, Volzhentsev A, Badmaev O, Luzan N, Lukyanenko K, Peters G, Lapin I, Kirichenko A, Konarev P, Morozov E, Mironov G, Gargaun A, Muharemagic D, Zamay S, Kochkina E, Dymova M, Smolyarova T, Sokolov A, Modestov A, Tokarev N, Shepelevich N, Ozerskaya A, Chanchikova N, Krat A, Zukov R, Bakhtina V, Shnyakin P, Shesternya P, Svetlichnyi V, Petrova M, Artyukhov I, Tomilin F, Berezovski M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
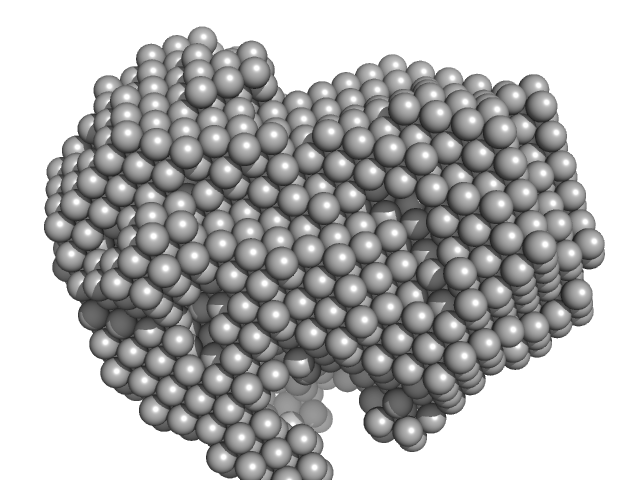
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
34 |
nm3 |
|
|