|
|
|
|
|
| Sample: |
Nucleoprotein dimer, 55 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
5.2 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF tetramer, 223 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jul 7
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
412 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF monomer, 25 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2023 Jan 18
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
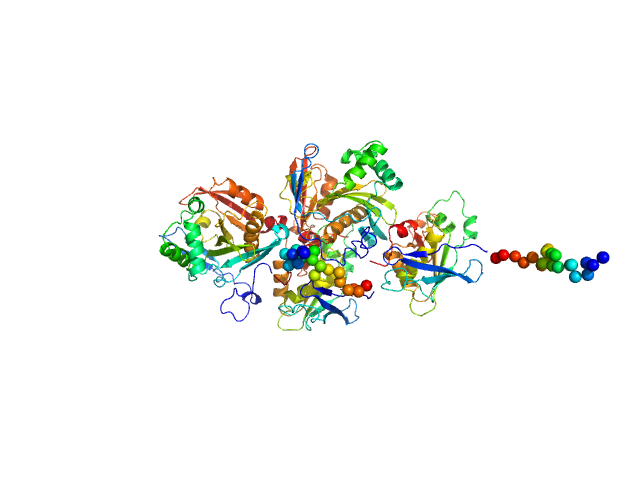
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF (Δ444-462) dimer, 107 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2023 Jan 18
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
559 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.0 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
546 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
563 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C3H1-type domain-containing protein monomer, 8 kDa Selaginella moellendorffii protein
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 9
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C3H1-type domain-containing protein monomer, 8 kDa Selaginella moellendorffii protein
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 9
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C3H1-type domain-containing protein monomer, 8 kDa Selaginella moellendorffii protein
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 9
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
14 |
nm3 |
|
|