|
|
|
|
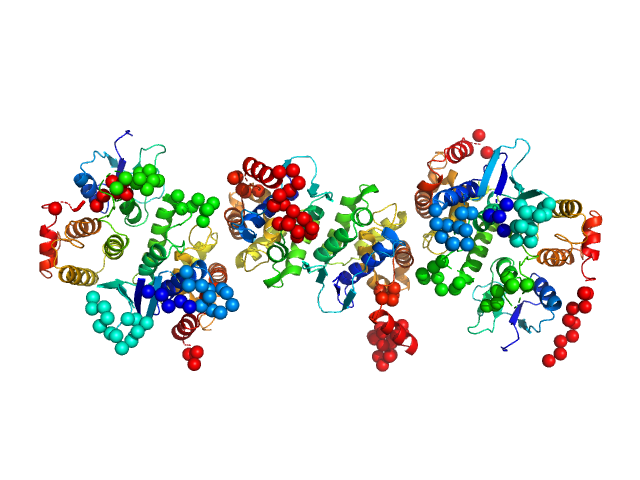
|
| Sample: |
Glutamine--tRNA ligase dimer, 46 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
tRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|
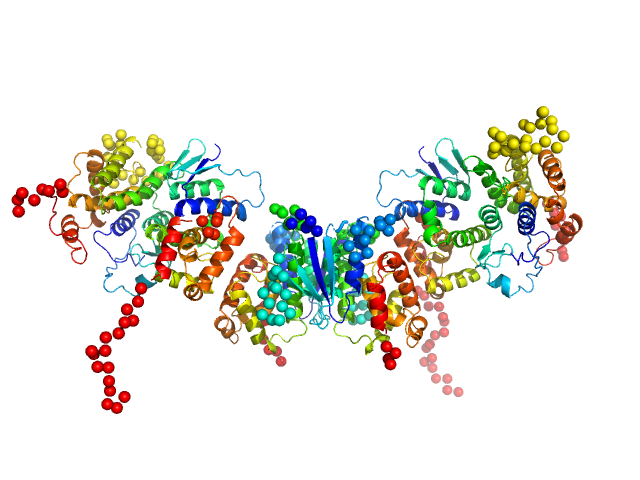
|
| Sample: |
Methionine--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
tRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dihydroneopterin aldolase tetramer, 55 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5 and 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Apr 5
|
Structure of Helicobacter pylori dihydroneopterin aldolase suggests a fragment-based strategy for isozyme-specific inhibitor design.
Curr Res Struct Biol 5:100095 (2023)
Shaw GX, Fan L, Cherry S, Shi G, Tropea JE, Ji X
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
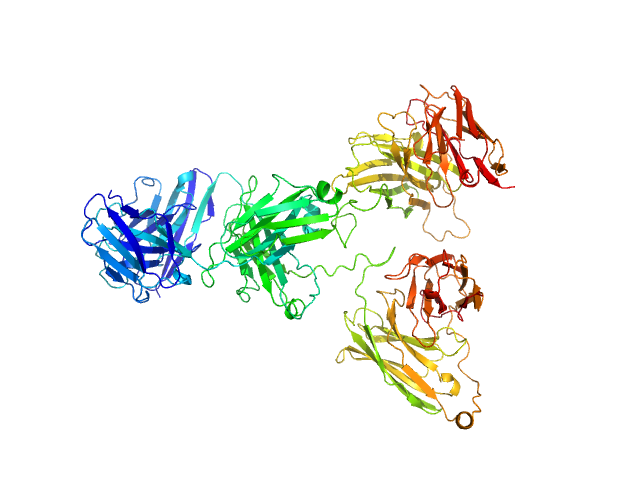
|
| Sample: |
Tri-specific antibody monomer, 102 kDa Homo sapiens protein
|
| Buffer: |
PBS, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 26
|
TRYBE®: an Fc-free antibody format with three monovalent targeting arms engineered for long in vivo half-life.
MAbs 15(1):2160229 (2023)
Davé E, Durrant O, Dhami N, Compson J, Broadbridge J, Archer S, Maroof A, Whale K, Menochet K, Bonnaillie P, Barry E, Wild G, Peerboom C, Bhatta P, Ellis M, Hinchliffe M, Humphreys DP, Heywood SP
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
High affinity immunoglobulin gamma Fc recombinant CD64 monomer, 32 kDa synthetic construct protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Dec 9
|
The solution structure of the unbound IgG Fc receptor CD64 resembles its crystal structure: Implications for function.
PLoS One 18(9):e0288351 (2023)
Hui GK, Gao X, Gor J, Lu J, Sun PD, Perkins SJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res (2022)
Keil P, Wulf A, Kachariya N, Reuscher S, Hühn K, Silbern I, Altmüller J, Keller M, Stehle R, Zarnack K, Sattler M, Urlaub H, Sträßer K
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res (2022)
Keil P, Wulf A, Kachariya N, Reuscher S, Hühn K, Silbern I, Altmüller J, Keller M, Stehle R, Zarnack K, Sattler M, Urlaub H, Sträßer K
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
Short RNA oligonucleotide (NPL3 binding sequence) monomer, 4 kDa RNA
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res (2022)
Keil P, Wulf A, Kachariya N, Reuscher S, Hühn K, Silbern I, Altmüller J, Keller M, Stehle R, Zarnack K, Sattler M, Urlaub H, Sträßer K
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein V monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
3.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein V (Y211A, Y212A, Y213A mutant) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|