|
|
|
|
|
| Sample: |
Bacteriorhodopsin monomer, 27 kDa Halobacterium salinarum protein
|
| Buffer: |
25 mM NaH2PO4, 1.35 mM KOH, 40 mM partially-deuterated octyl glucoside mixture, pH: 5.6
|
| Experiment: |
SANS
data collected at NGB 30m SANS, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2017 Jan 20
|
Direct localization of detergents and bacteriorhodopsin in the lipidic cubic phase by small-angle neutron scattering
IUCrJ 8(1) (2021)
Cleveland IV T, Blick E, Krueger S, Leung A, Darwish T, Butler P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bacteriorhodopsin monomer, 27 kDa Halobacterium salinarum protein
|
| Buffer: |
25 mM NaH2PO4, 1.35 mM KOH, 40 mM octyl glucoside, pH: 5.6
|
| Experiment: |
SANS
data collected at NG7, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2017 May 15
|
Direct localization of detergents and bacteriorhodopsin in the lipidic cubic phase by small-angle neutron scattering
IUCrJ 8(1) (2021)
Cleveland IV T, Blick E, Krueger S, Leung A, Darwish T, Butler P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
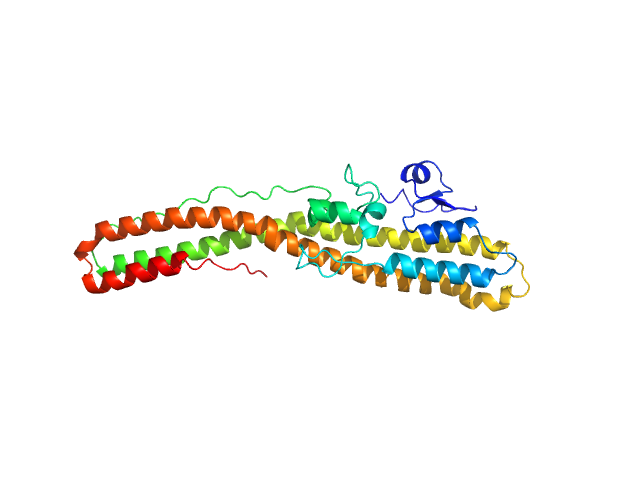
| Sample: |
Tetanus toxin (C467S) monomer, 46 kDa Clostridium tetani protein
|
| Buffer: |
10 mM HEPES 100 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL45XU, SPring-8 on 2018 Oct 20
|
Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X 5:100045 (2021)
Zhang CM, Imoto Y, Hikima T, Inoue T
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tetanus toxin monomer, 35 kDa Clostridium tetani protein
|
| Buffer: |
10 mM HEPES 100 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL45XU, SPring-8 on 2018 Oct 20
|
Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X 5:100045 (2021)
Zhang CM, Imoto Y, Hikima T, Inoue T
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
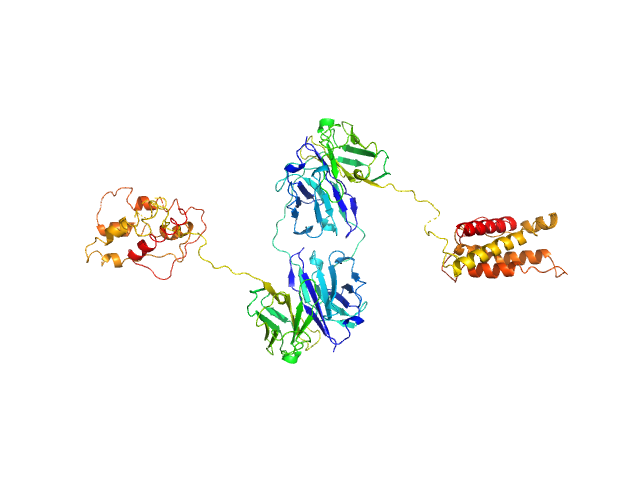
| Sample: |
L19-IL2 dimeric immunocytokine dimer, 84 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES/NaOH, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 23
|
Inference of molecular structure for characterization and improvement of clinical grade immunocytokines
Journal of Structural Biology :107696 (2021)
Ongaro T, Guarino S, Scietti L, Palamini M, Wulhfard S, Neri D, Villa A, Forneris F
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
L19L19-IL2 immunocytokine monomer, 69 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES/NaOH, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Mar 4
|
Inference of molecular structure for characterization and improvement of clinical grade immunocytokines
Journal of Structural Biology :107696 (2021)
Ongaro T, Guarino S, Scietti L, Palamini M, Wulhfard S, Neri D, Villa A, Forneris F
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
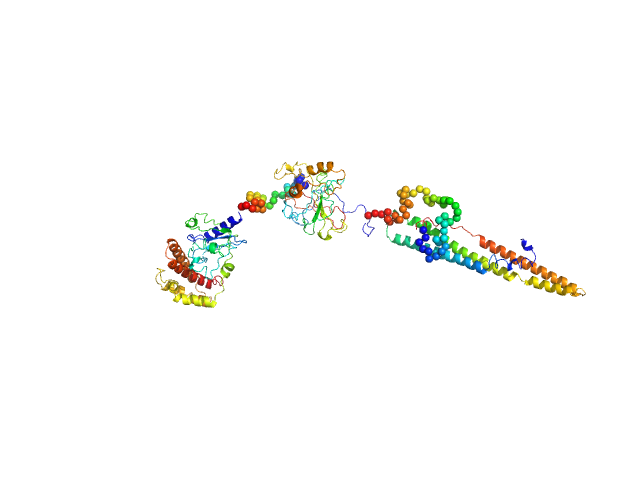
| Sample: |
IL12-L19L19 immunocytokine monomer, 112 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES/NaOH, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Mar 4
|
Inference of molecular structure for characterization and improvement of clinical grade immunocytokines
Journal of Structural Biology :107696 (2021)
Ongaro T, Guarino S, Scietti L, Palamini M, Wulhfard S, Neri D, Villa A, Forneris F
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 27 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 20
|
Structural Analysis of the Partially Disordered Protein EspK from Mycobacterium Tuberculosis
Crystals 11(1):18 (2020)
Gijsbers A, Sánchez-Puig N, Gao Y, Peters P, Ravelli R, Siliqi D
|
| RgGuinier |
7.2 |
nm |
| Dmax |
3.3 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 30 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 12
|
Structural Analysis of the Partially Disordered Protein EspK from Mycobacterium Tuberculosis
Crystals 11(1):18 (2020)
Gijsbers A, Sánchez-Puig N, Gao Y, Peters P, Ravelli R, Siliqi D
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Adagio protein 1 (Zeitlupe G46S:G80R) dimer, 37 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Nov 2
|
Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry (2020)
Pudasaini A, Green R, Song YH, Blumenfeld A, Karki N, Imaizumi T, Zoltowski BD
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|