|
|
|
|

|
| Sample: |
Transcription intermediary factor 1-beta dimer, 82 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris 300 mM NaCl 0.1 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Aug 10
|
A Dissection of Oligomerisation by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
J Mol Biol (2019)
Sun Y, Keown JR, Black MM, Raclot C, Demarais N, Trono D, Turelli P, Goldstone DC
|
| RgGuinier |
7.0 |
nm |
| Dmax |
23.2 |
nm |
| VolumePorod |
232 |
nm3 |
|
|
|
|
|
|
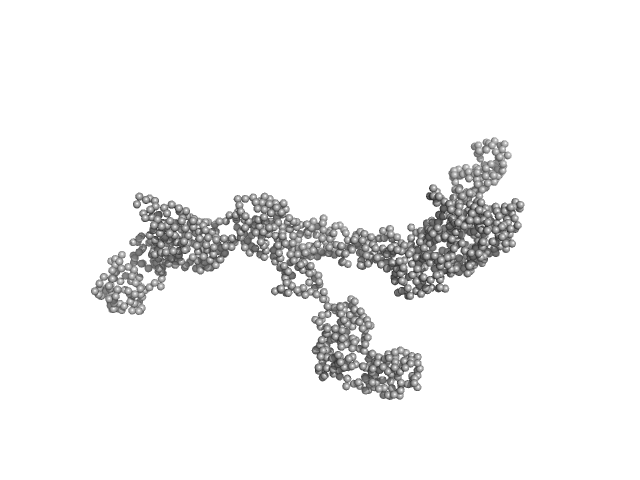
|
| Sample: |
Transcription intermediary factor 1-beta dimer, 82 kDa Mus musculus protein
Zinc finger protein 809 N-terminal MBP fusion monomer, 52 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris 300 mM NaCl 0.1 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Aug 10
|
A Dissection of Oligomerisation by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
J Mol Biol (2019)
Sun Y, Keown JR, Black MM, Raclot C, Demarais N, Trono D, Turelli P, Goldstone DC
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 51 kDa Rattus norvegicus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Apr 24
|
Structure, Function, and Dynamics of the Gα Binding Domain of Ric-8A.
Structure (2019)
Zeng B, Mou TC, Doukov TI, Steiner A, Yu W, Papasergi-Scott M, Tall GG, Hagn F, Sprang SR
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 51 kDa Rattus norvegicus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Apr 24
|
Structure, Function, and Dynamics of the Gα Binding Domain of Ric-8A.
Structure (2019)
Zeng B, Mou TC, Doukov TI, Steiner A, Yu W, Papasergi-Scott M, Tall GG, Hagn F, Sprang SR
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dosage compensation regulator monomer, 29 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM NaPO4, 200 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 29
|
Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless.
Nucleic Acids Res 47(8):4319-4333 (2019)
Ankush Jagtap PK, Müller M, Masiewicz P, von Bülow S, Hollmann NM, Chen PC, Simon B, Thomae AW, Becker PB, Hennig J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dosage compensation regulator monomer, 29 kDa Drosophila melanogaster protein
roX2 stem-loop 7, 18-mer fragment monomer, 12 kDa synthetic construct RNA
|
| Buffer: |
20 mM NaPO4, 200 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 29
|
Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless.
Nucleic Acids Res 47(8):4319-4333 (2019)
Ankush Jagtap PK, Müller M, Masiewicz P, von Bülow S, Hollmann NM, Chen PC, Simon B, Thomae AW, Becker PB, Hennig J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
roX2 stem-loop 7, 18-mer fragment monomer, 12 kDa synthetic construct RNA
|
| Buffer: |
20 mM NaPO4, 200 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 29
|
Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless.
Nucleic Acids Res 47(8):4319-4333 (2019)
Ankush Jagtap PK, Müller M, Masiewicz P, von Bülow S, Hollmann NM, Chen PC, Simon B, Thomae AW, Becker PB, Hennig J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
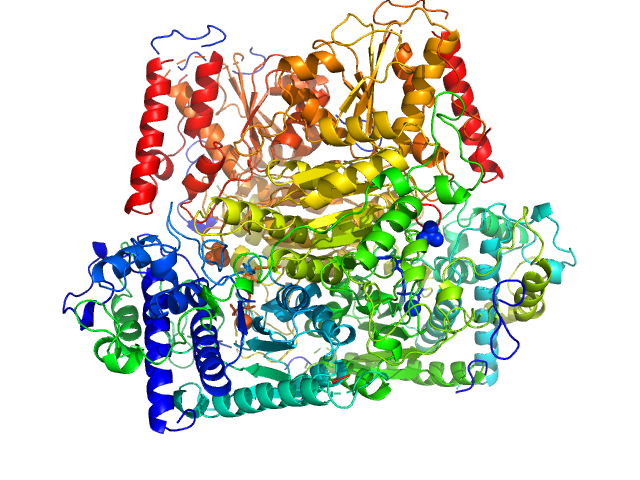
|
| Sample: |
Probable phosphoketolase dimer, 191 kDa Lactococcus lactis subsp. … protein
|
| Buffer: |
20mM potassium phosphate 150mM NaCl 0.007 %(w/v) β-octyl glucoside 1mM DTT 1mM MgCl 1mM thiaminpyrophosphate 2 %(v/v) glycerol, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 7
|
Crystal structure of a xylulose 5-phosphate phosphoketolase. insights into the substrate specificity for xylulose 5-phosphate.
J Struct Biol (2019)
Scheidig AJ, Horvath D, Szedlacsek SE
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
|
|
|
|
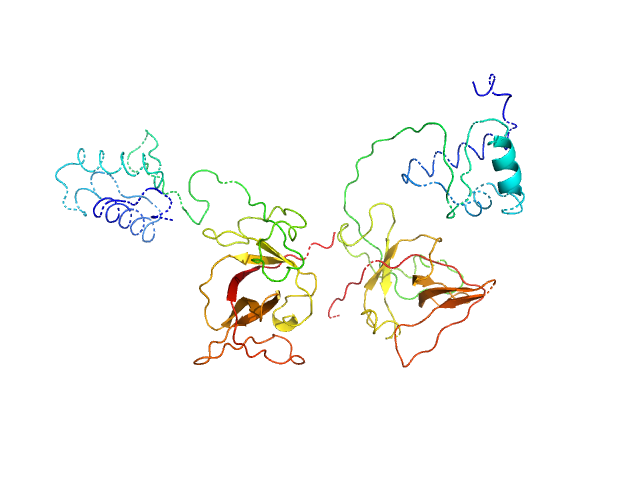
|
| Sample: |
Bacillus thuringiensis LexA repressor dimer, 47 kDa Bacillus thuringiensis protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 10% glycerol,, pH: 8
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Aug 25
|
Structural Insights into Bacteriophage GIL01 gp7 Inhibition of Host LexA Repressor.
Structure 27(7):1094-1102.e4 (2019)
Caveney NA, Pavlin A, Caballero G, Bahun M, Hodnik V, de Castro L, Fornelos N, Butala M, Strynadka NCJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
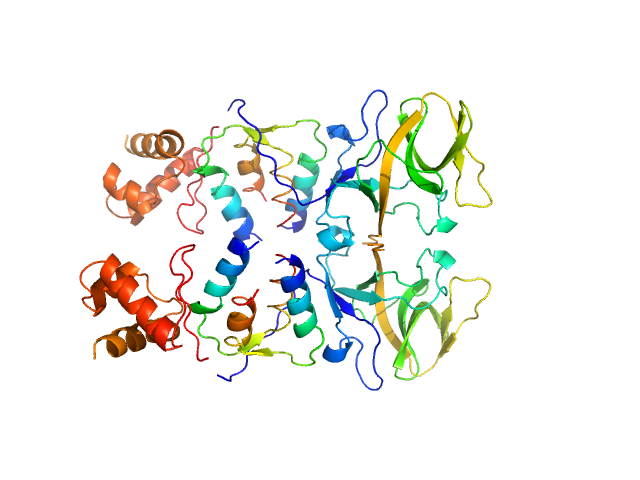
|
| Sample: |
Bacillus thuringiensis LexA repressor dimer, 47 kDa Bacillus thuringiensis protein
Bacteriophage pGIL01 gp7 tetramer, 24 kDa Bacteriophage pGIL01 protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 10% glycerol,, pH: 8
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Aug 25
|
Structural Insights into Bacteriophage GIL01 gp7 Inhibition of Host LexA Repressor.
Structure 27(7):1094-1102.e4 (2019)
Caveney NA, Pavlin A, Caballero G, Bahun M, Hodnik V, de Castro L, Fornelos N, Butala M, Strynadka NCJ
|
|
|