|
|
|
|
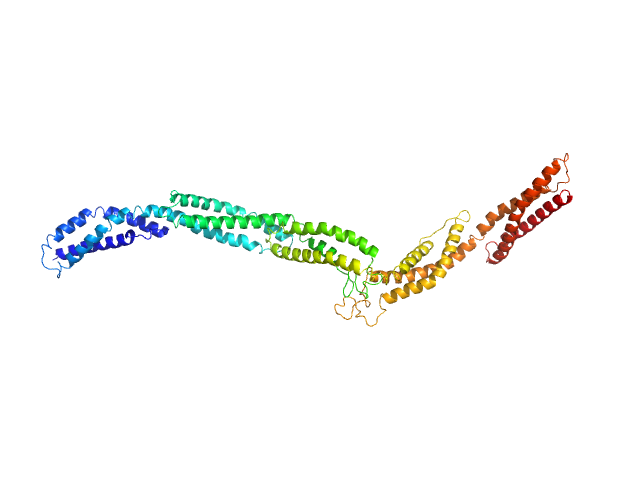
|
| Sample: |
Dystrophin central domain repeats 20 to 24. monomer, 67 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Jul 10
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F...
|
| RgGuinier |
5.8 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
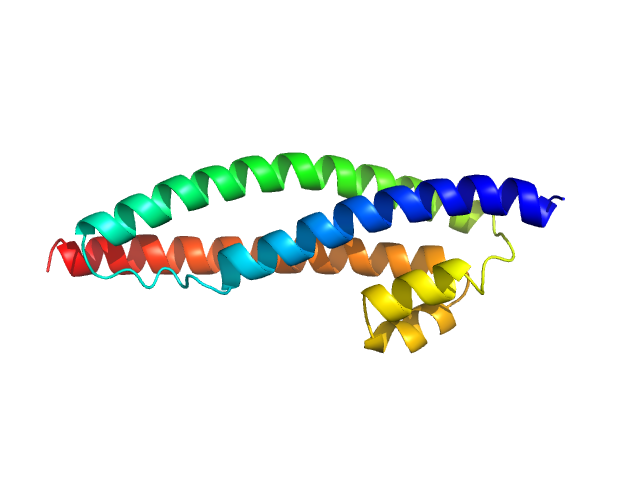
|
| Sample: |
Dystrophin central domain single repeat 23 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Oct 7
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F...
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
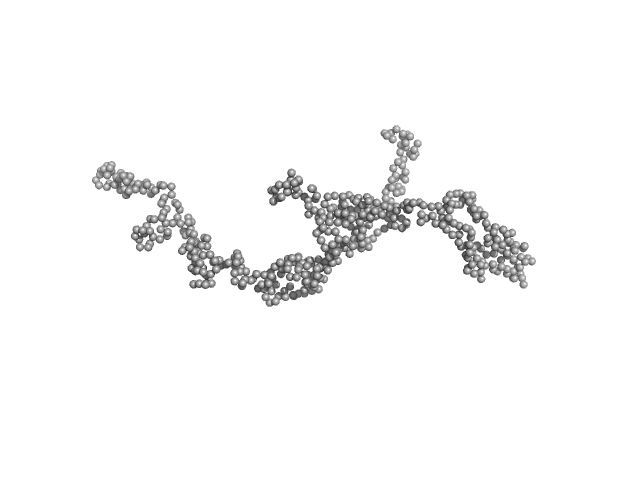
|
| Sample: |
Dystrophin central domain repeats 16 to 21 (Δ2146-2305; Becker muscular dystrophy variant, deletion of exons 45-47) monomer, 64 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol 5% acetonitrile, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Feb 5
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F...
|
| RgGuinier |
6.0 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
184 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fluorescence recovery protein dimer, 23 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.1 mM EDTA, 2 mM dithiothreitol, 3 % v/v glycerol, pH: 7.6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
Functional interaction of low-homology FRPs from different cyanobacteria with Synechocystis OCP.
Biochim Biophys Acta 1859(5):382-393 (2018)
Slonimskiy YB, Maksimov EG, Lukashev EP, Moldenhauer M, Jeffries CM, Svergun DI, Friedrich T, Sluchanko NN
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Uncharacterized fluorescence recovery protein dimer, 24 kDa Arthrospira maxima CS-328 protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.1 mM EDTA, 2 mM dithiothreitol, 3 % v/v glycerol, pH: 7.6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
Functional interaction of low-homology FRPs from different cyanobacteria with Synechocystis OCP.
Biochim Biophys Acta 1859(5):382-393 (2018)
Slonimskiy YB, Maksimov EG, Lukashev EP, Moldenhauer M, Jeffries CM, Svergun DI, Friedrich T, Sluchanko NN
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
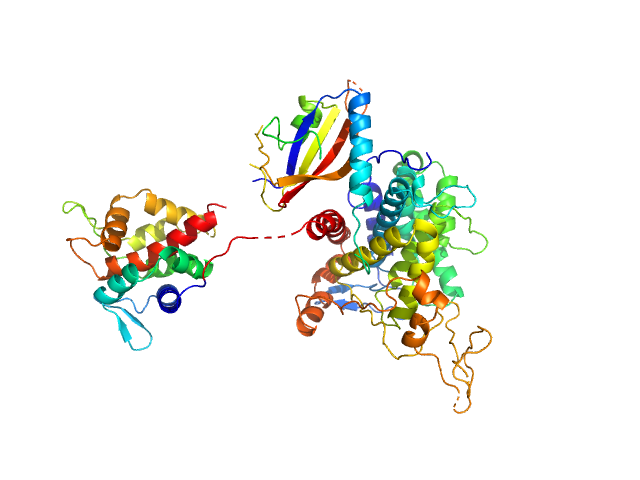
|
| Sample: |
Ubiquitinating/deubiquitinating enzyme SdeA monomer, 72 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 26
|
Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination.
Nature 557(7707):734-738 (2018)
Kalayil S, Bhogaraju S, Bonn F, Shin D, Liu Y, Gan N, Basquin J, Grumati P, Luo ZQ, Dikic I
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
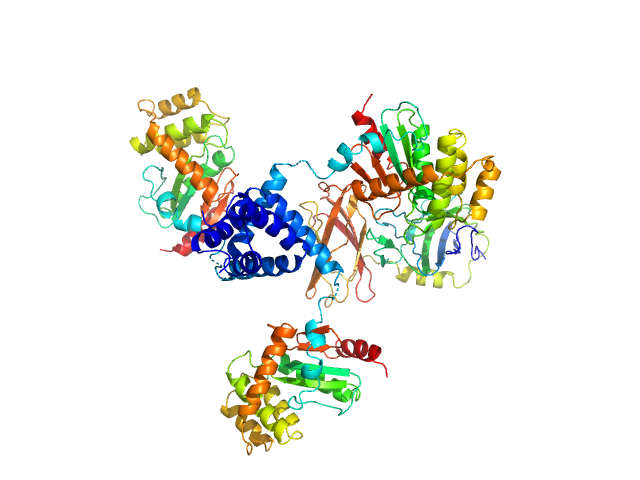
DsbA-like protein trimer, 74 kDa Proteus mirabilis protein
Putative metal resistance protein monomer, 30 kDa Proteus mirabilis protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 2
|
Disulfide isomerase activity of the dynamic, trimeric Proteus mirabilis ScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J Biol Chem 293(16):5793-5805 (2018)
Furlong EJ, Choudhury HG, Kurth F, Duff AP, Whitten AE, Martin JL
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
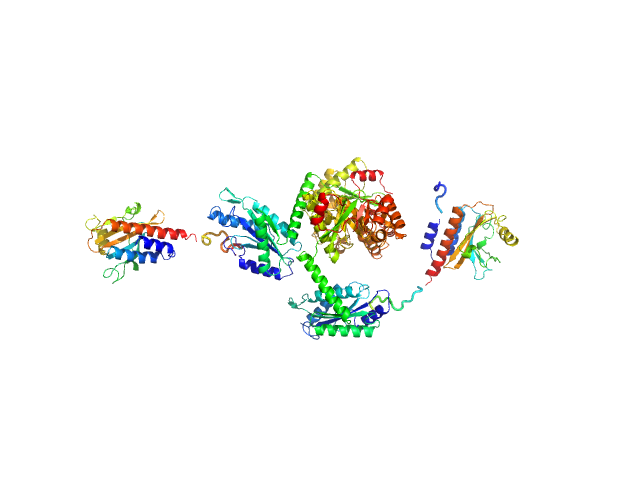
Sensory box/response regulator dimer, 136 kDa Mycobacterium smegmatis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5% glycerol, 2 mM MgCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jun 26
|
The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP in Mycobacterium smegmatis.
Biochem J 475(7):1295-1308 (2018)
Chen HJ, Li N, Luo Y, Jiang YL, Zhou CZ, Chen Y, Li Q
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
299 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
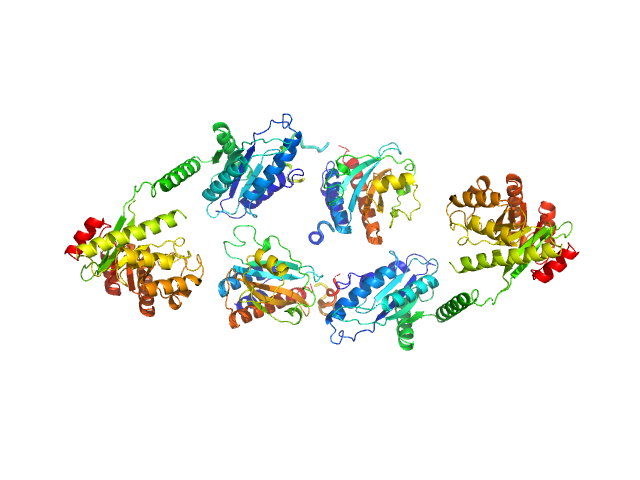
Sensory box/response regulator dimer, 136 kDa Mycobacterium smegmatis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5% glycerol, 2 mM MgCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jun 26
|
The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP in Mycobacterium smegmatis.
Biochem J 475(7):1295-1308 (2018)
Chen HJ, Li N, Luo Y, Jiang YL, Zhou CZ, Chen Y, Li Q
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
271 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calvin cycle protein CP12, chloroplastic monomer, 11 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
50 mM phosphate buffer, 50 mM NaCl, 20 mM oxidized DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 3
|
Cryptic Disorder Out of Disorder: Encounter between Conditionally Disordered CP12 and Glyceraldehyde-3-Phosphate Dehydrogenase.
J Mol Biol 430(8):1218-1234 (2018)
Launay H, Barré P, Puppo C, Zhang Y, Maneville S, Gontero B, Receveur-Bréchot V
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|