|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 processively polymerizes length-controlled matriglycan on prodystroglycan.
Nat Commun 16(1):9028 (2025)
Joseph S, Schnicker NJ, Spellmon N, Xu Z, Yan R, Yu Z, Davulcu O, Yang T, Hopkins J, Anderson ME, Venzke D, Campbell KP
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
264 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 processively polymerizes length-controlled matriglycan on prodystroglycan.
Nat Commun 16(1):9028 (2025)
Joseph S, Schnicker NJ, Spellmon N, Xu Z, Yan R, Yu Z, Davulcu O, Yang T, Hopkins J, Anderson ME, Venzke D, Campbell KP
|
| RgGuinier |
4.3 |
nm |
| Dmax |
18.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 processively polymerizes length-controlled matriglycan on prodystroglycan.
Nat Commun 16(1):9028 (2025)
Joseph S, Schnicker NJ, Spellmon N, Xu Z, Yan R, Yu Z, Davulcu O, Yang T, Hopkins J, Anderson ME, Venzke D, Campbell KP
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.5 |
nm |
|
|
|
|
|
|
|
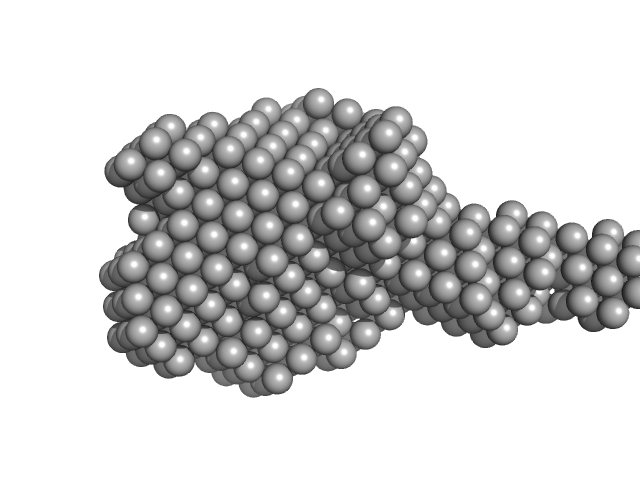
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 processively polymerizes length-controlled matriglycan on prodystroglycan.
Nat Commun 16(1):9028 (2025)
Joseph S, Schnicker NJ, Spellmon N, Xu Z, Yan R, Yu Z, Davulcu O, Yang T, Hopkins J, Anderson ME, Venzke D, Campbell KP
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
231 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Structural polyprotein dimer, 26 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 27
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Structural polyprotein (Capsid protein VP3: K947R; Δ756-843; Δ977-1012) monomer, 18 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM TRIS, 500 mM NaCl, 2 mM DTT,, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Structural polyprotein (Capsid protein VP3: K947R; Δ977-1012) dimer, 55 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM TRIS, 500 mM NaCl, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 1
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|