|
|
|
|
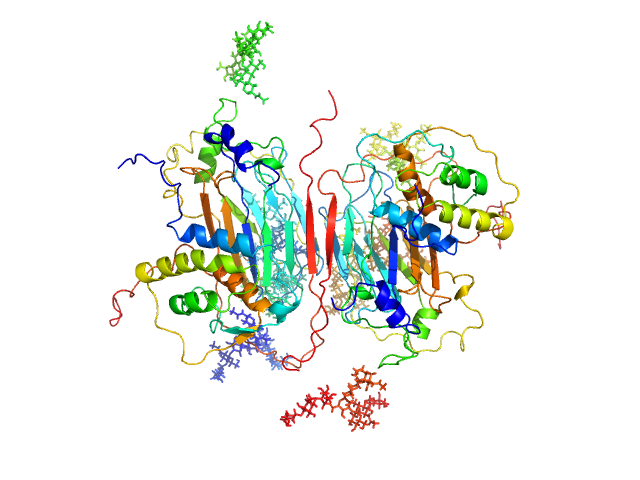
|
| Sample: |
GM-CSF/IL-2 inhibition factor tetramer, 120 kDa Orf virus protein
Granulocyte-macrophage colony-stimulating factor dimer, 29 kDa Ovis aries protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Sep 10
|
Structural basis of GM-CSF and IL-2 sequestration by the viral decoy receptor GIF.
Nat Commun 7:13228 (2016)
Felix J, Kandiah E, De Munck S, Bloch Y, van Zundert GC, Pauwels K, Dansercoer A, Novanska K, Read RJ, Bonvin AM, Vergauwen B, Verstraete K, Gutsche I, Savvides SN
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
231 |
nm3 |
|