UniProt ID: P21333 (2151-2329) Human Filamin A Ig-like domains 20-21
|
|
|
|
| Sample: |
Human Filamin A Ig-like domains 20-21 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 1
|
Flexible Structure of Peptide-Bound Filamin A Mechanosensor Domain Pair 20-21.
PLoS One 10(8):e0136969 (2015)
Seppälä J, Tossavainen H, Rodic N, Permi P, Pentikäinen U, Ylänne J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: P21333 (2141-2329) Human Filamin A Ig-like domains 20-21*/migfilin peptide complex
|
|
|
|
| Sample: |
Human Filamin A Ig-like domains 20-21*/migfilin peptide complex monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 1
|
Flexible Structure of Peptide-Bound Filamin A Mechanosensor Domain Pair 20-21.
PLoS One 10(8):e0136969 (2015)
Seppälä J, Tossavainen H, Rodic N, Permi P, Pentikäinen U, Ylänne J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: P21333 (2141-2329) Human Filamin A Ig-like domains 20-21*
|
|
|

|
| Sample: |
Human Filamin A Ig-like domains 20-21* monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 1
|
Flexible Structure of Peptide-Bound Filamin A Mechanosensor Domain Pair 20-21.
PLoS One 10(8):e0136969 (2015)
Seppälä J, Tossavainen H, Rodic N, Permi P, Pentikäinen U, Ylänne J
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: O41974 (124-316) Latency-associated nuclear antigen
|
|
|
|
| Sample: |
Latency-associated nuclear antigen tetramer, 87 kDa Murid herpesvirus 4 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
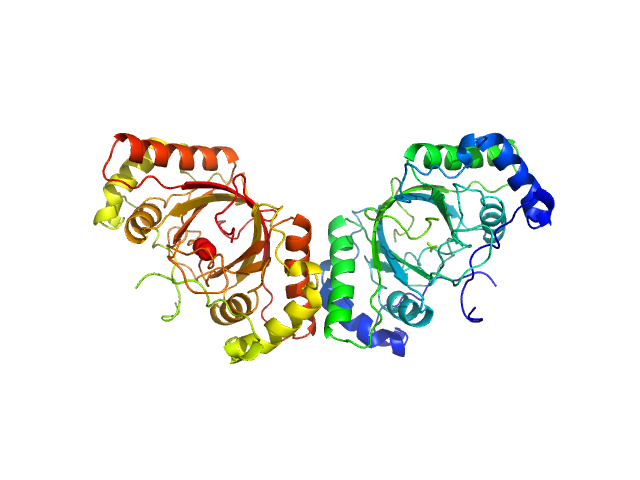
UniProt ID: Q9QR71 (1008-1150) ORF73 tetramer
UniProt ID: Q9QR71 (1008-1150) ORF73 dimer
|
|
|
|
| Sample: |
ORF73 tetramer tetramer, 63 kDa Human herpesvirus 8 protein
ORF73 dimer dimer, 32 kDa Human herpesvirus 8 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
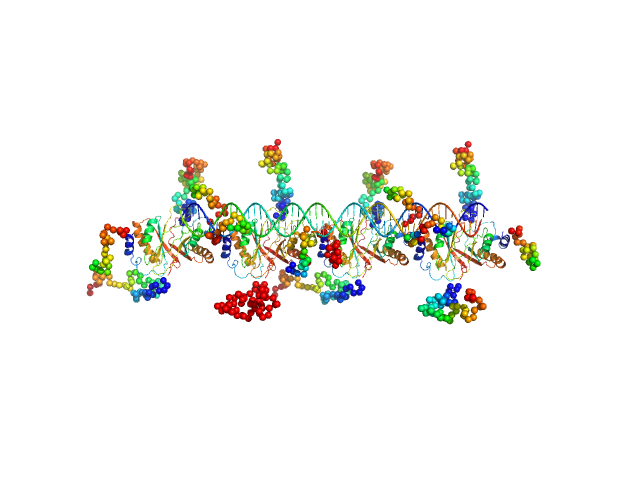
UniProt ID: (None-None) MHV-68 TR DNA
UniProt ID: O41974 (124-316) Latency-associated nuclear antigen
|
|
|
|
| Sample: |
MHV-68 TR DNA monomer, 30 kDa unidentified herpesvirus DNA
Latency-associated nuclear antigen octamer, 269 kDa Murid herpesvirus 4 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
5.8 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
475 |
nm3 |
|
|
UniProt ID: (None-None) kLBS1-2 DNA
UniProt ID: Q9QR71 (1008-1150) ORF73 tetramer
UniProt ID: Q9QR71 (1008-1150) ORF73 octamer
UniProt ID: (None-None) kLBS1-2 DNA two monomers
|
|
|

|
| Sample: |
KLBS1-2 DNA monomer, 24 kDa unidentified herpesvirus DNA
ORF73 tetramer tetramer, 63 kDa Human herpesvirus 8 protein
ORF73 octamer octamer, 126 kDa Human herpesvirus 8 protein
KLBS1-2 DNA two monomers dimer, 48 kDa unidentified herpesvirus RNA
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
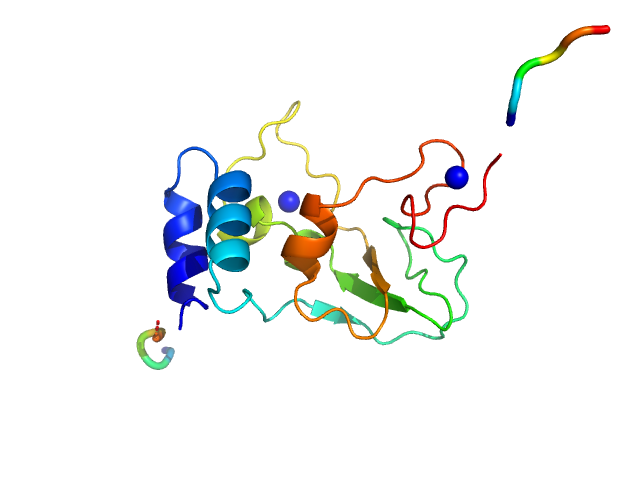
UniProt ID: P0C6U2 (3934-4068) HCoV-229E Non-structural protein 10
|
|
|
|
| Sample: |
HCoV-229E Non-structural protein 10 monomer, 15 kDa Human coronavirus 229E protein
|
| Buffer: |
25 mM HEPES 280 mM NaCl 2 mM DTT 500 µM ZnCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 25
|
Human alphacoronavirus non-structural protein Nsp10
Sven Falke,
Al Kikhney
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.8 |
nm |
|
|
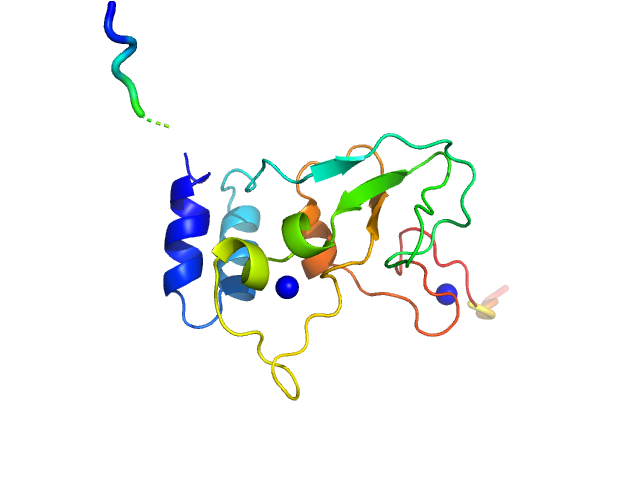
UniProt ID: P0C6U2 (3934-4068) HCoV-229E Non-structural protein 10
|
|
|
|
| Sample: |
HCoV-229E Non-structural protein 10 monomer, 15 kDa Human coronavirus 229E protein
|
| Buffer: |
25 mM HEPES 400 mM NaCl 1 mM EDTA 5% glycerol 40 mM NaH2PO4, pH: 7.9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2011 Sep 8
|
Human alphacoronavirus non-structural protein Nsp10
Sven Falke,
Al Kikhney
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.9 |
nm |
|
|
UniProt ID: Q9J5U5 (None-None) GM-CSF/IL-2 inhibition factor
UniProt ID: P28773 (18-144) Granulocyte-macrophage colony-stimulating factor
|
|
|
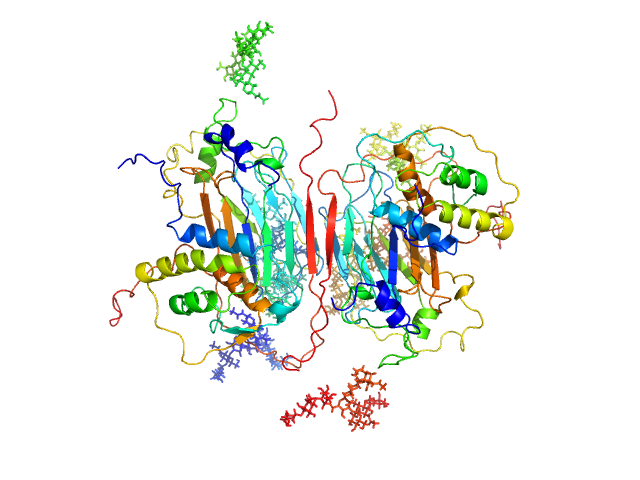
|
| Sample: |
GM-CSF/IL-2 inhibition factor tetramer, 120 kDa Orf virus protein
Granulocyte-macrophage colony-stimulating factor dimer, 29 kDa Ovis aries protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Sep 10
|
Structural basis of GM-CSF and IL-2 sequestration by the viral decoy receptor GIF.
Nat Commun 7:13228 (2016)
Felix J, Kandiah E, De Munck S, Bloch Y, van Zundert GC, Pauwels K, Dansercoer A, Novanska K, Read RJ, Bonvin AM, Vergauwen B, Verstraete K, Gutsche I, Savvides SN
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
231 |
nm3 |
|
|