UniProt ID: P40424 (1-308) Pre-B-cell leukemia transcription factor 1
UniProt ID: P55347 (1-344) Homeobox protein PKNOX1
|
|
|
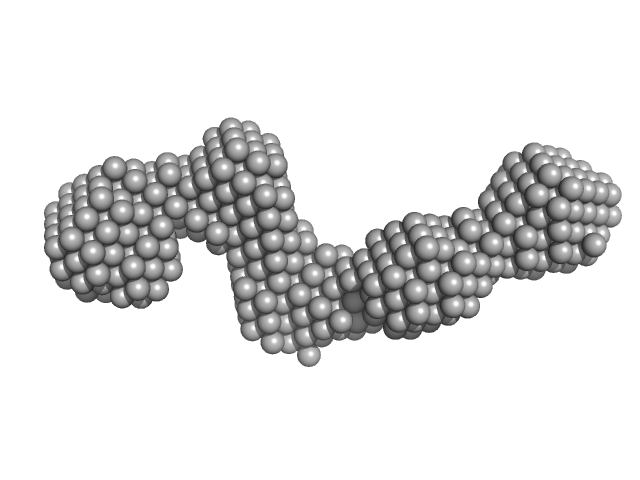
|
| Sample: |
Pre-B-cell leukemia transcription factor 1 monomer, 34 kDa Homo sapiens protein
Homeobox protein PKNOX1 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
UniProt ID: P40424 (1-308) Pre-B-cell leukemia transcription factor 1
UniProt ID: P55347 (1-344) Homeobox protein PKNOX1
UniProt ID: (None-None) DNA 22mer
|
|
|
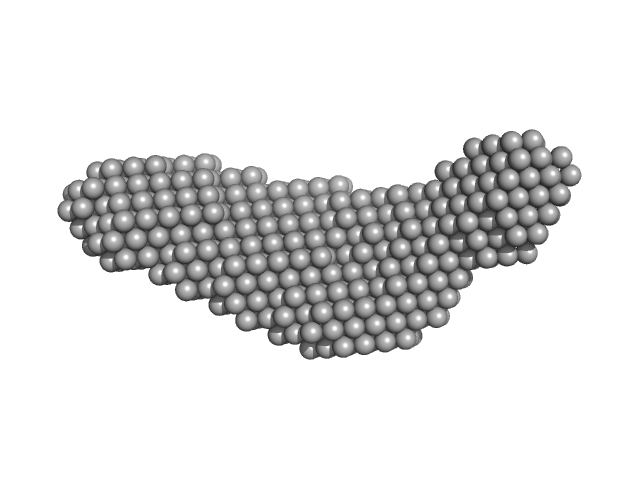
|
| Sample: |
Pre-B-cell leukemia transcription factor 1 monomer, 34 kDa Homo sapiens protein
Homeobox protein PKNOX1 monomer, 38 kDa Homo sapiens protein
DNA 22mer monomer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
UniProt ID: O88998 (None-None) Noelin
|
|
|
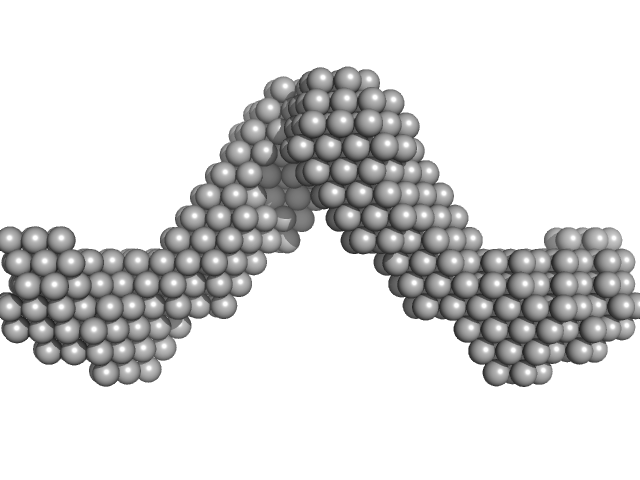
|
| Sample: |
Noelin tetramer, 256 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 6
|
Olfactomedin-1 Has a V-shaped Disulfide-linked Tetrameric Structure.
J Biol Chem 290(24):15092-101 (2015)
Pronker MF, Bos TG, Sharp TH, Thies-Weesie DM, Janssen BJ
|
| RgGuinier |
8.5 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
616 |
nm3 |
|
|
UniProt ID: P08887 (None-None) Interleukin-6 receptor subunit alpha
UniProt ID: P08887 (None-None) Interleukin-6 receptor subunit alpha
|
|
|
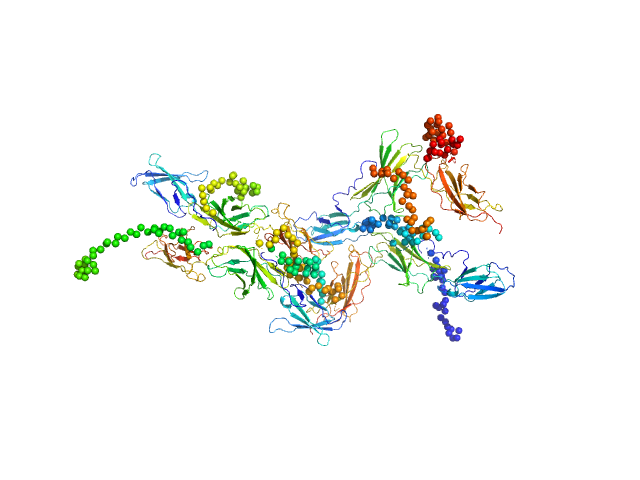
|
| Sample: |
Interleukin-6 receptor subunit alpha tetramer, 164 kDa Homo sapiens protein
Interleukin-6 receptor subunit alpha dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2014 Oct 1
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
4.9 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
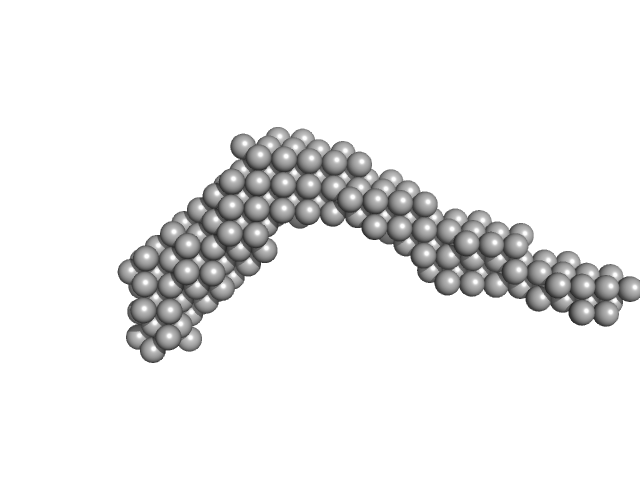
UniProt ID: (None-None) AIR-3A
UniProt ID: P08887 (None-None) Interleukin-6 receptor subunit alpha
|
|
|
|
| Sample: |
AIR-3A tetramer, 26 kDa RNA
Interleukin-6 receptor subunit alpha dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2014 Oct 1
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
6.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
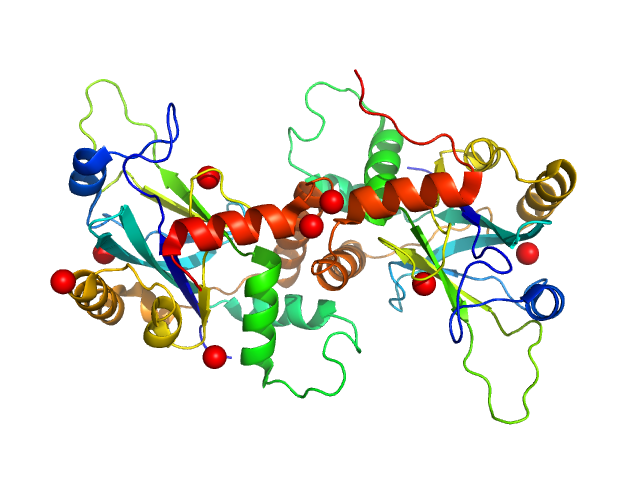
UniProt ID: Q83417 (1-224) VP24
|
|
|
|
| Sample: |
VP24 dimer, 53 kDa Suid herpesvirus 1 protein
|
| Buffer: |
50 mM Tris/HCl 0.5 M NaCl 0.25 M imidazole 5% glycerol 50 mM urea 0.2 M MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease.
PLoS Pathog 11(7):e1005045 (2015)
Zühlsdorf M, Werten S, Klupp BG, Palm GJ, Mettenleiter TC, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
UniProt ID: Q9EP80 (2-416) PRKCA-binding protein
|
|
|
|
| Sample: |
PRKCA-binding protein dimer, 93 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM Tris 125 mM NaCl 0.01 vol% reduced TX-100, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2015 May 11
|
Structure of Dimeric and Tetrameric Complexes of the BAR Domain Protein PICK1 Determined by Small-Angle X-Ray Scattering.
Structure 23(7):1258-1270 (2015)
Karlsen ML, Thorsen TS, Johner N, Ammendrup-Johnsen I, Erlendsson S, Tian X, Simonsen JB, Høiberg-Nielsen R, Christensen NM, Khelashvili G, Streicher W, Teilum K, Vestergaard B, Weinstein H, Gether U, Arleth L, Madsen KL
|
| RgGuinier |
6.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
205 |
nm3 |
|
|
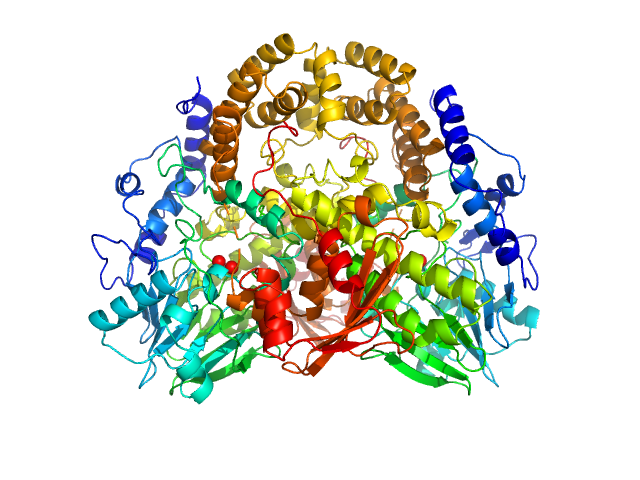
UniProt ID: Q9I5I9 (None-None) Pseudomonas aeruginosa SDS hydrolase SdsA1
|
|
|
|
| Sample: |
Pseudomonas aeruginosa SDS hydrolase SdsA1 dimer, 145 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
50 mM HEPES, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2013 Jul 22
|
SdsA polymorph isolation and improvement of their crystal quality using nonconventional crystallization techniques
Journal of Applied Crystallography 48(5):1551-1559 (2015)
De la Mora E, Flores-Hernández E, Jakoncic J, Stojanoff V, Siliqi D, Sánchez-Puig N, Moreno A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
261 |
nm3 |
|
|
UniProt ID: Q08FF8 (1-155) Antiapoptotic membrane protein
|
|
|
|
| Sample: |
Antiapoptotic membrane protein dimer, 39 kDa Deerpox virus W-1170-84 protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Structural basis of Deerpox virus-mediated inhibition of apoptosis.
Acta Crystallogr D Biol Crystallogr 71(Pt 8):1593-603 (2015)
Burton DR, Caria S, Marshall B, Barry M, Kvansakul M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
UniProt ID: P21333 (2151-2329) Human Filamin A Ig-like domains 20-21/migfilin peptide complex
|
|
|
|
| Sample: |
Human Filamin A Ig-like domains 20-21/migfilin peptide complex monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 1
|
Flexible Structure of Peptide-Bound Filamin A Mechanosensor Domain Pair 20-21.
PLoS One 10(8):e0136969 (2015)
Seppälä J, Tossavainen H, Rodic N, Permi P, Pentikäinen U, Ylänne J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
31 |
nm3 |
|
|