UniProt ID: Q04207 (19-549) Transcription factor p65 19-549
UniProt ID: P25799 (39-350) NF-kappa-B p105 subunit 39-350
UniProt ID: None (None-None) Urokinase kB
|
|
|
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Urokinase kB monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 4
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
UniProt ID: P05090 (21-189) Apolipoprotein D
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
UniProt ID: P05090 (21-189) Apolipoprotein D
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
164 |
nm3 |
|
|
UniProt ID: P05090 (21-189) Apolipoprotein D
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
UniProt ID: P05090 (21-189) Apolipoprotein D
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
UniProt ID: P05090 (21-189) Apolipoprotein D
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
171 |
nm3 |
|
|
UniProt ID: A0A0H3B5N9 (1-247) Regulator Of Virulence interconnected with the Csr system
|
|
|
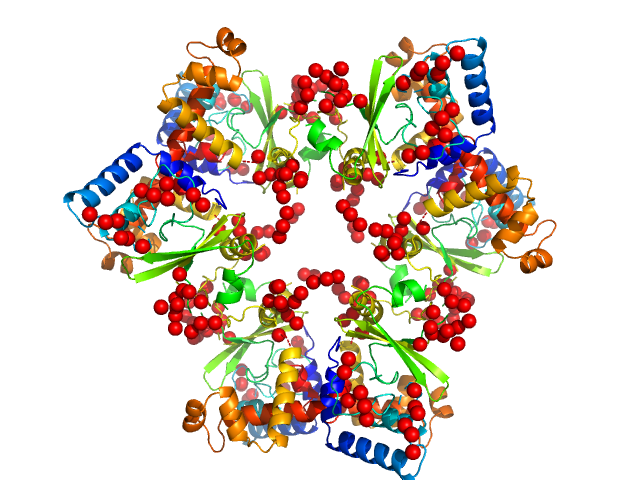
|
| Sample: |
Regulator Of Virulence interconnected with the Csr system hexamer, 175 kDa Yersinia pseudotuberculosis protein
|
| Buffer: |
50 mM TRIS pH= 8, 500 mM NaCl, 5 mM DTT, 5 % v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 18
|
RovC - a novel type of hexameric transcriptional activator promoting type VI secretion gene expression
PLOS Pathogens 16(9):e1008552 (2020)
Knittel V, Sadana P, Seekircher S, Stolle A, Körner B, Volk M, Jeffries C, Svergun D, Heroven A, Scrima A, Dersch P, Mecsas J
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
UniProt ID: Q793I8 (1-184) TRAF-interacting protein with FHA domain-containing protein A
|
|
|

|
| Sample: |
TRAF-interacting protein with FHA domain-containing protein A dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 100 mM arginine, 5 % glycerol, 10 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Kumamoto University on 2014 Oct 16
|
Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep 10(1):5152 (2020)
Nakamura T, Hashikawa C, Okabe K, Yokote Y, Chirifu M, Toma-Fukai S, Nakamura N, Matsuo M, Kamikariya M, Okamoto Y, Gohda J, Akiyama T, Semba K, Ikemizu S, Otsuka M, Inoue JI, Yamagata Y
|
| RgGuinier |
3.1 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
UniProt ID: Q16790 (38-414) Carbonic anhydrase 9
|
|
|
|
| Sample: |
Carbonic anhydrase 9 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Nov 16
|
Biophysical Characterization of Cancer-Related Carbonic Anhydrase IX.
Int J Mol Sci 21(15) (2020)
Koruza K, Murray AB, Mahon BP, Hopkins JB, Knecht W, McKenna R, Fisher SZ
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
UniProt ID: P49189 (2-494) 4-trimethylaminobutyraldehyde dehydrogenase
|
|
|
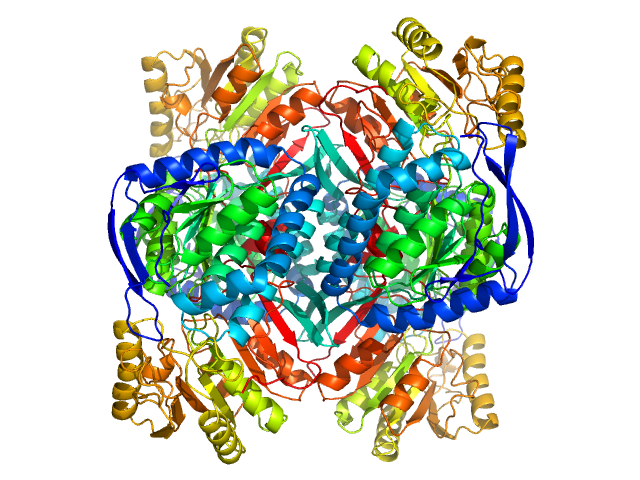
|
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
252 |
nm3 |
|
|