UniProt ID: Q15366 (1-365) Poly(rC)-binding protein 2
UniProt ID: None (None-None) modified stem loop IV poliovirus IRES, nucleotides 278-398
|
|
|
|
| Sample: |
Poly(rC)-binding protein 2 monomer, 40 kDa Homo sapiens protein
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 16
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
UniProt ID: None (None-None) modified stem loop IV poliovirus IRES, nucleotides 278-398
UniProt ID: Q15366 (1-253) Truncated poly(rC)-binding protein 2 (ΔKH3)
UniProt ID: Q15366 (1-253) Truncated poly(rC)-binding protein 2 (ΔKH3)
|
|
|
|
| Sample: |
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 1
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
UniProt ID: Q15366 (1-253) Truncated poly(rC)-binding protein 2 (ΔKH3)
|
|
|
|
| Sample: |
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jan 1
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
UniProt ID: None (None-None) modified stem loop IV poliovirus IRES, nucleotides 278-398
UniProt ID: Q15366 (11-168) Truncated poly(rC)-binding protein 2 (ΔKH1-KH2)
|
|
|
|
| Sample: |
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
Truncated poly(rC)-binding protein 2 (ΔKH1-KH2) monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 1
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
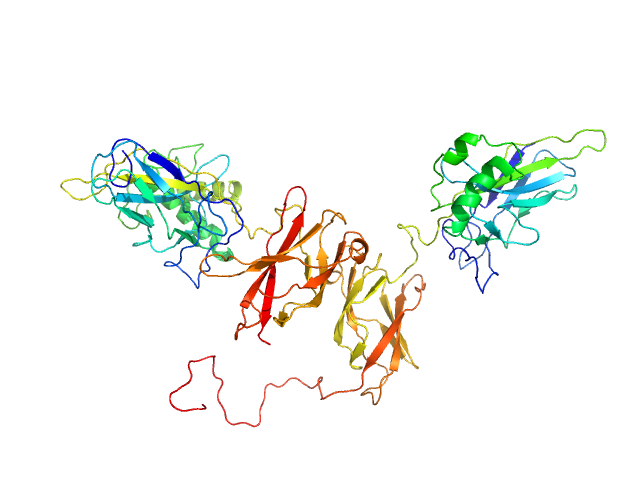
UniProt ID: P25799 (39-350) NF-kappa-B p105 subunit 39-350
UniProt ID: Q04207 (19-321) Transcription factor p65 19-321
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
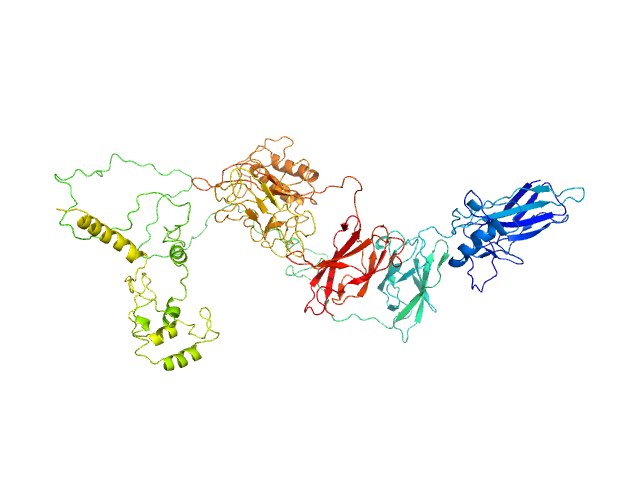
UniProt ID: Q04207 (19-549) Transcription factor p65 19-549
UniProt ID: P25799 (39-350) NF-kappa-B p105 subunit 39-350
|
|
|
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
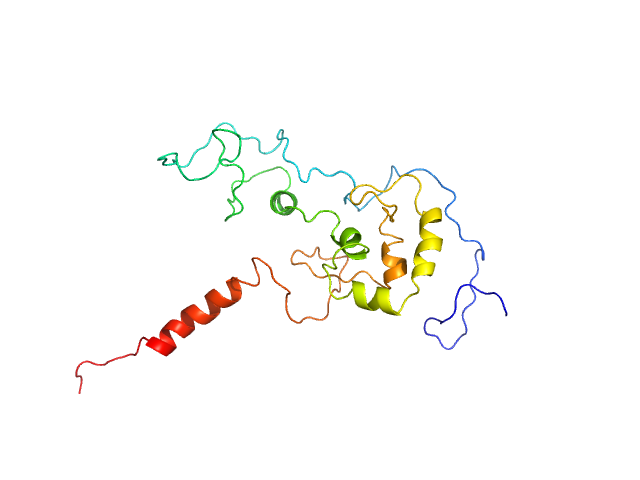
UniProt ID: Q04207 (340-549) Transcription factor p65 340-549
|
|
|
|
| Sample: |
Transcription factor p65 340-549 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
UniProt ID: P25799 (39-350) NF-kappa-B p105 subunit 39-350
UniProt ID: Q04207 (19-321) Transcription factor p65 19-321
UniProt ID: None (None-None) IFN kB DNA
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
IFN kB DNA monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
UniProt ID: P25799 (39-350) NF-kappa-B p105 subunit 39-350
UniProt ID: Q04207 (19-321) Transcription factor p65 19-321
UniProt ID: None (None-None) Urokinase kB
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
Urokinase kB monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
UniProt ID: Q04207 (19-549) Transcription factor p65 19-549
UniProt ID: P25799 (39-350) NF-kappa-B p105 subunit 39-350
UniProt ID: None (None-None) IFN kB DNA
|
|
|
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
IFN kB DNA monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 4
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
314 |
nm3 |
|
|