UniProt ID: A0A1X9WII3 (1-113) Emfourin (Protealysin-associated protein)
|
|
|

|
| Sample: |
Emfourin (Protealysin-associated protein) monomer, 13 kDa Serratia proteamaculans protein
|
| Buffer: |
50 mM Tris-HCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Apr 22
|
NMR structure of emfourin, a novel protein metalloprotease inhibitor: insights into the mechanism of action.
J Biol Chem :104585 (2023)
Bozin T, Berdyshev I, Chukhontseva K, Karaseva M, Konarev P, Varizhuk A, Lesovoy D, Arseniev A, Kostrov S, Bocharov E, Demidyuk I
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
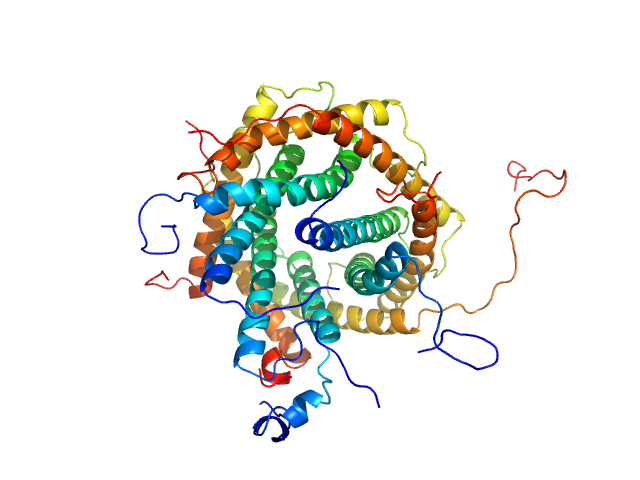
UniProt ID: P32830 (2-109) Mitochondrial import inner membrane translocase subunit TIM12
UniProt ID: O74700 (1-87) Mitochondrial import inner membrane translocase subunit TIM9
UniProt ID: P87108 (2-93) Mitochondrial import inner membrane translocase subunit TIM10
|
|
|
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM12 monomer, 12 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM9 dimer, 20 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 trimer, 31 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
20 mM MES buffer (2-(N-morpholino)ethanesulfonic acid), pH 6.5, 50 mM NaCl Cloning,, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 8
|
Architecture and subunit dynamics of the mitochondrial TIM9·10·12 chaperone
(2020)
Weinhäupl K, Wang Y, Hessel A, Brennich M, Lindorff-Larsen K, Schanda P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
UniProt ID: P15516 (20-43) Histatin-3, His3-(20-43)-peptide
|
|
|
|
| Sample: |
Histatin-3, His3-(20-43)-peptide monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 13
|
Comment on the Optimal Parameters to Derive Intrinsically Disordered Protein Conformational Ensembles from Small-Angle X-ray Scattering Data Using the Ensemble Optimization Method
Journal of Chemical Theory and Computation (2021)
Sagar A, Jeffries C, Petoukhov M, Svergun D, Bernadó P
|
| RgGuinier |
1.4 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
UniProt ID: P15516 (20-43) Histatin-3, His3-(20-43)-peptide
|
|
|
|
| Sample: |
Histatin-3, His3-(20-43)-peptide monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 13
|
Comment on the Optimal Parameters to Derive Intrinsically Disordered Protein Conformational Ensembles from Small-Angle X-ray Scattering Data Using the Ensemble Optimization Method
Journal of Chemical Theory and Computation (2021)
Sagar A, Jeffries C, Petoukhov M, Svergun D, Bernadó P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
UniProt ID: P15516 (20-43) Histatin-3, His3-(20-43)-peptide
|
|
|
|
| Sample: |
Histatin-3, His3-(20-43)-peptide monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 13
|
Comment on the Optimal Parameters to Derive Intrinsically Disordered Protein Conformational Ensembles from Small-Angle X-ray Scattering Data Using the Ensemble Optimization Method
Journal of Chemical Theory and Computation (2021)
Sagar A, Jeffries C, Petoukhov M, Svergun D, Bernadó P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
UniProt ID: P9WHG9 (660-738) ACT domain of Rel protein (Bifunctional (p)ppGpp synthase/hydrolase RelA)
|
|
|
|
| Sample: |
ACT domain of Rel protein (Bifunctional (p)ppGpp synthase/hydrolase RelA) dimer, 20 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 350 mM NaCl, 5% glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, Nanyang Technological University on 2018 Jun 7
|
Atomic structure of, and valine binding to the regulatory ACT domain of the Mycobacterium tuberculosis Rel protein.
FEBS J (2020)
Shin J, Singal B, Manimekalai MSS, Chen MW, Ragunathan P, Grüber G
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
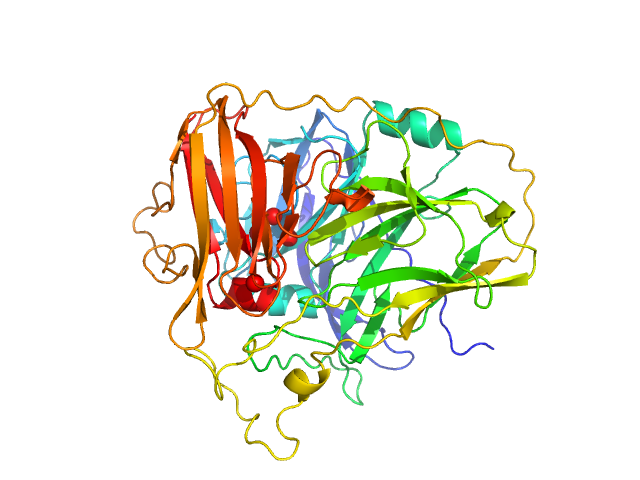
UniProt ID: O67206 (44-527) McoA evolved variant 2F4 (Periplasmic cell division protein (SufI))
|
|
|
|
| Sample: |
McoA evolved variant 2F4 (Periplasmic cell division protein (SufI)) monomer, 55 kDa Aquifex aeolicus VF5 protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 25
|
Distal Mutations Shape Substrate-Binding Sites during Evolution of a Metallo-Oxidase into a Laccase
ACS Catalysis :5022-5035 (2022)
Brissos V, Borges P, Núñez-Franco R, Lucas M, Frazão C, Monza E, Masgrau L, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
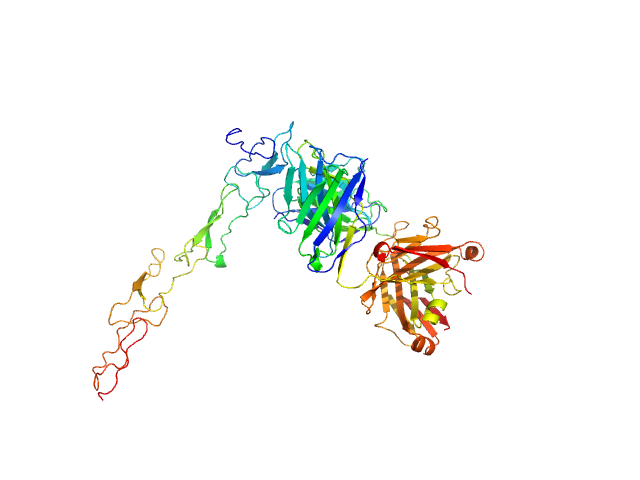
UniProt ID: P25942 (21-193) Tumor necrosis factor receptor superfamily member 5
UniProt ID: None (None-None) Human 341G2 F(ab)
|
|
|
|
| Sample: |
Tumor necrosis factor receptor superfamily member 5 monomer, 19 kDa Homo sapiens protein
Human 341G2 F(ab) monomer, 73 kDa Homo sapiens protein
|
| Buffer: |
Phosphate-buffered saline, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
Isotype Switching Converts Anti-CD40 Antagonism to Agonism to Elicit Potent Antitumor Activity
Cancer Cell (2020)
Yu X, Chan H, Fisher H, Penfold C, Kim J, Inzhelevskaya T, Mockridge C, French R, Duriez P, Douglas L, English V, Verbeek J, White A, Tews I, Glennie M, Cragg M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
UniProt ID: G0S061 (1-1581) Pentafunctional AROM polypeptide
|
|
|
|
| Sample: |
Pentafunctional AROM polypeptide dimer, 345 kDa Chaetomium thermophilum protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl. 2 mM TCEP, 1% sucrose, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Nov 22
|
Architecture and functional dynamics of the pentafunctional AROM complex
Nature Chemical Biology (2020)
Arora Verasztó H, Logotheti M, Albrecht R, Leitner A, Zhu H, Hartmann M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
623 |
nm3 |
|
|
UniProt ID: F1BV96 (1-2084) Severe fever with thrombocytopenia syndrome virus L protein - RNA-dependent RNA polymerase
|
|
|
|
| Sample: |
Severe fever with thrombocytopenia syndrome virus L protein - RNA-dependent RNA polymerase monomer, 238 kDa SFTS virus AH12 protein
|
| Buffer: |
50 mM HEPES(NaOH) pH 7.0, 500 mM NaCl, 5% (w/v) glycerol, and 2 mM dithiothreitol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 13
|
Structural and functional characterization of the Severe fever with thrombocytopenia syndrome virus L protein
(2020)
Vogel D, Thorkelsson S, Quemin E, Meier K, Kouba T, Gogrefe N, Busch C, Reindl S, Günther S, Cusack S, Grünewald K, Rosenthal M
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
483 |
nm3 |
|
|