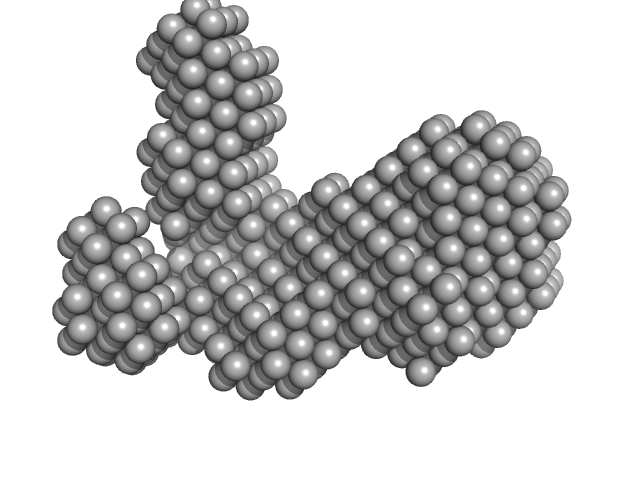
UniProt ID: P9WMQ1 (1-771) ATP-dependent DNA helicase UvrD1
|
|
|
|
| Sample: |
ATP-dependent DNA helicase UvrD1 monomer, 85 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
10 mM Tris-Cl, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2020 Feb 28
|
Structural and biochemical characterization of RNA polymerase core and UvrD complex: a key component in transcription coupled DNA repair
Ravishankar Ramachandran
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
UniProt ID: P0CL43 (27-147) Type 3 secretion system pilotin
|
|
|

|
| Sample: |
Type 3 secretion system pilotin dimer, 28 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Mar 23
|
Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure (2020)
Majewski DD, Okon M, Heinkel F, Robb CS, Vuckovic M, McIntosh LP, Strynadka NCJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: P0CL43 (70-147) Type 3 secretion system pilotin
|
|
|

|
| Sample: |
Type 3 secretion system pilotin dimer, 19 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Mar 23
|
Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure (2020)
Majewski DD, Okon M, Heinkel F, Robb CS, Vuckovic M, McIntosh LP, Strynadka NCJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: G2JBB2 (2-291) Candidatus Glomeribacter gigasporarum cyclodipeptide synthase
UniProt ID: None (None-None) E. coli Phe-tRNAPhe
|
|
|
|
| Sample: |
Candidatus Glomeribacter gigasporarum cyclodipeptide synthase monomer, 34 kDa Candidatus Glomeribacter gigasporarum protein
E. coli Phe-tRNAPhe monomer, 25 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS pH6.7; 200 mM NaCl, 8 mM MgCl2, pH: 6.7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Oct 2
|
Structural basis of the interaction between cyclodipeptide synthases and aminoacylated tRNA substrates.
RNA 26(11):1589-1602 (2020)
Bourgeois G, Seguin J, Babin M, Gondry M, Mechulam Y, Schmitt E
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
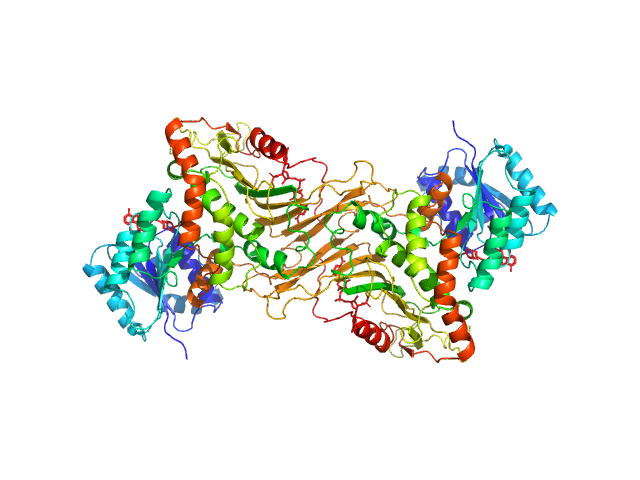
UniProt ID: P11413 (1-515) Glucose-6-phosphate 1-dehydrogenase
|
|
|
|
| Sample: |
Glucose-6-phosphate 1-dehydrogenase dimer, 119 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 24
|
Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
Proceedings of the National Academy of Sciences 118(4) (2021)
Horikoshi N, Hwang S, Gati C, Matsui T, Castillo-Orellana C, Raub A, Garcia A, Jabbarpour F, Batyuk A, Broweleit J, Xiang X, Chiang A, Broweleit R, Vöhringer-Martinez E, Mochly-Rosen D, Wakatsuki S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
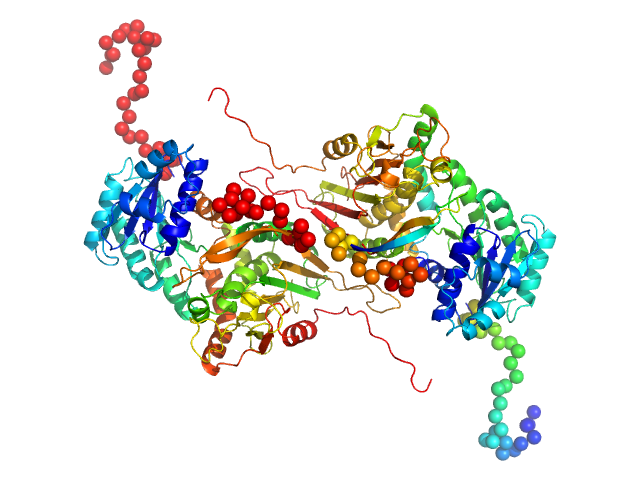
UniProt ID: P11413 (1-515) Glucose-6-phosphate 1-dehydrogenase P396L
|
|
|
|
| Sample: |
Glucose-6-phosphate 1-dehydrogenase P396L dimer, 119 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 24
|
Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
Proceedings of the National Academy of Sciences 118(4) (2021)
Horikoshi N, Hwang S, Gati C, Matsui T, Castillo-Orellana C, Raub A, Garcia A, Jabbarpour F, Batyuk A, Broweleit J, Xiang X, Chiang A, Broweleit R, Vöhringer-Martinez E, Mochly-Rosen D, Wakatsuki S
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
178 |
nm3 |
|
|
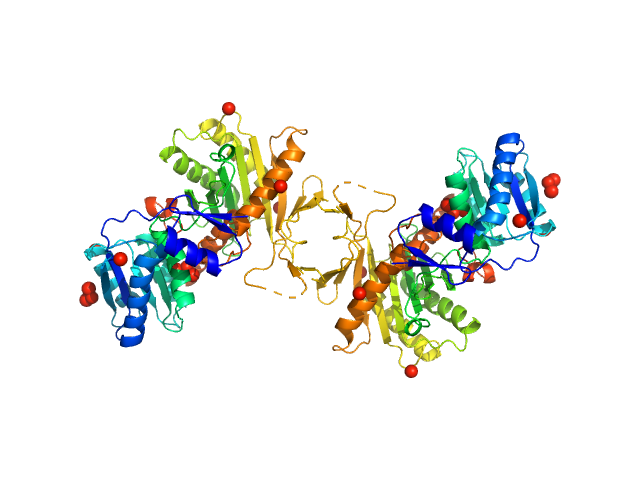
UniProt ID: O28951 (1-427) Piwi protein AF_1318
|
|
|
|
| Sample: |
Piwi protein AF_1318 dimer, 98 kDa Archaeoglobus fulgidus protein
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 500 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
Apo Archaeoglobus fulgidus Argonaute protein dimerises in solution
Elena Manakova
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
174 |
nm3 |
|
|
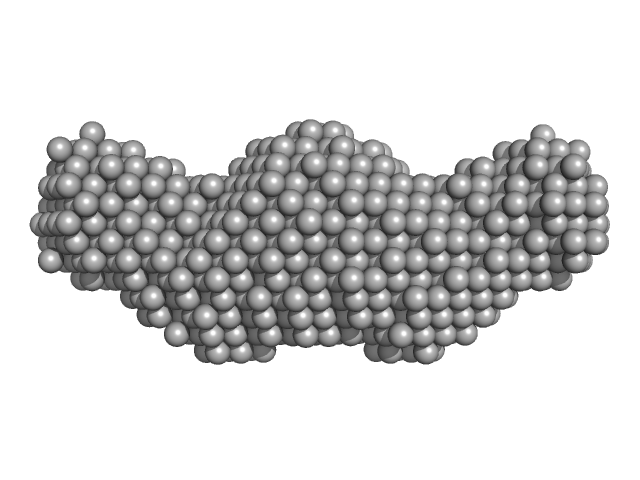
UniProt ID: Q9Y6M5 (342-507) Zinc transporter 1
|
|
|
|
| Sample: |
Zinc transporter 1 dimer, 37 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 14
|
Heterologous Expression and Biochemical Characterization of the Human Zinc Transporter 1 (ZnT1) and Its Soluble C-Terminal Domain
Frontiers in Chemistry 9 (2021)
Cotrim C, Jarrott R, Whitten A, Choudhury H, Drew D, Martin J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
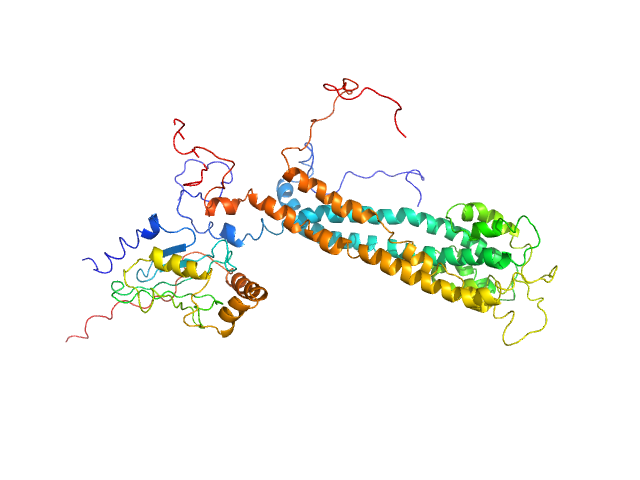
UniProt ID: Q07457 (1-212) E3 ubiquitin-protein ligase BRE1
UniProt ID: P06104 (1-172) Ubiquitin-conjugating enzyme E2 2
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase BRE1 dimer, 50 kDa Saccharomyces cerevisiae (strain … protein
Ubiquitin-conjugating enzyme E2 2 monomer, 20 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Feb 11
|
Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones
International Journal of Biological Macromolecules :127717 (2023)
Yadav P, Gupta M, Wazahat R, Islam Z, Tsutakawa S, Kamthan M, Kumar P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
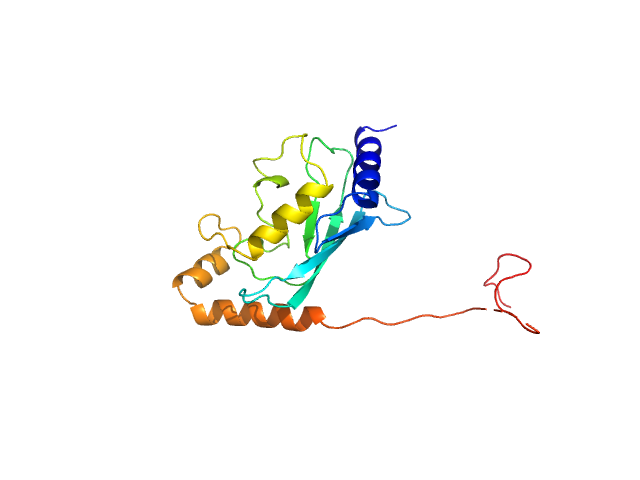
UniProt ID: P06104 (1-172) Ubiquitin-conjugating enzyme E2 2
|
|
|
|
| Sample: |
Ubiquitin-conjugating enzyme E2 2 monomer, 20 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Feb 11
|
Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones
International Journal of Biological Macromolecules :127717 (2023)
Yadav P, Gupta M, Wazahat R, Islam Z, Tsutakawa S, Kamthan M, Kumar P
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
31 |
nm3 |
|
|