UniProt ID: Q9IK91 (1-406) Phosphoprotein
|
|
|
|
| Sample: |
Phosphoprotein monomer, 45 kDa Nipah henipavirus protein
|
| Buffer: |
20 mM Tris-HCl, 0.3 M NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 3
|
Ensemble description of the intrinsically disordered N-terminal domain of the Nipah virus P/V protein from combined NMR and SAXS.
Sci Rep 10(1):19574 (2020)
Schiavina M, Salladini E, Murrali MG, Tria G, Felli IC, Pierattelli R, Longhi S
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
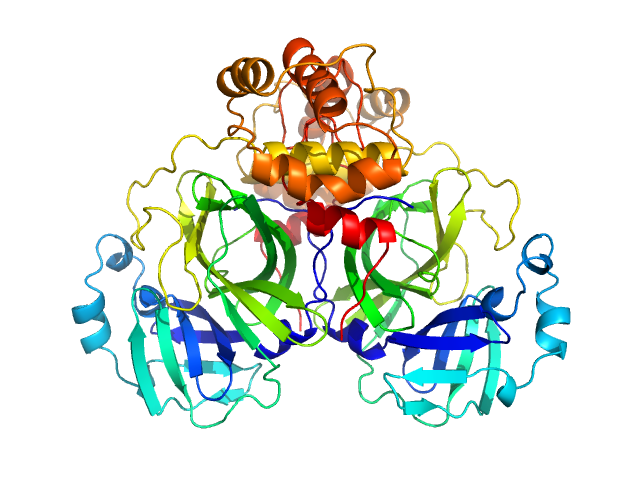
UniProt ID: P0DTC1 (3264-3569) 3C-like proteinase from SARS-CoV-2 replicase polyprotein 1a
|
|
|
|
| Sample: |
3C-like proteinase from SARS-CoV-2 replicase polyprotein 1a dimer, 68 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 1 mM DTT, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2020 Jun 1
|
Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun 11(1):5877 (2020)
Lee J, Worrall LJ, Vuckovic M, Rosell FI, Gentile F, Ton AT, Caveney NA, Ban F, Cherkasov A, Paetzel M, Strynadka NCJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
UniProt ID: Q8IW19 (449-511) Aprataxin and PNK-like factor (acidic domain)
UniProt ID: P84051 (2-124) Histone H2A
UniProt ID: P02283 (2-123) Histone H2B
UniProt ID: P02299 (2-136) Histone H3
UniProt ID: P84040 (2-103) Histone H4
|
|
|
|
| Sample: |
Aprataxin and PNK-like factor (acidic domain) dimer, 15 kDa Homo sapiens protein
Histone H2A dimer, 26 kDa Drosophila melanogaster protein
Histone H2B dimer, 27 kDa Drosophila melanogaster protein
Histone H3 dimer, 30 kDa Drosophila melanogaster protein
Histone H4 dimer, 23 kDa Drosophila melanogaster protein
|
| Buffer: |
25 mM NaPi, 300 mM NaCl, 3% v/v glycerol, 1 mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 10
|
Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv 8(30):eabo0517 (2022)
Corbeski I, Guo X, Eckhardt BV, Fasci D, Wiegant W, Graewert MA, Vreeken K, Wienk H, Svergun DI, Heck AJR, van Attikum H, Boelens R, Sixma TK, Mattiroli F, van Ingen H
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
UniProt ID: P31483-2 (93-274) Nucleolysin TIA-1 isoform p40
UniProt ID: None (None-None) TC1
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
TC1 monomer, 3 kDa synthetic construct DNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Jun 26
|
Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1.
Nucleic Acids Res (2021)
Loughlin FE, West DL, Gunzburg MJ, Waris S, Crawford SA, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
UniProt ID: P31483-2 (93-274) Nucleolysin TIA-1 isoform p40
UniProt ID: None (None-None) UC1
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
UC1 monomer, 3 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Jun 26
|
Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1.
Nucleic Acids Res (2021)
Loughlin FE, West DL, Gunzburg MJ, Waris S, Crawford SA, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
UniProt ID: P04958 (459-863) Tetanus toxin (C467S)
|
|
|
|
| Sample: |
Tetanus toxin (C467S) monomer, 46 kDa Clostridium tetani protein
|
| Buffer: |
10 mM HEPES 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL45XU, SPring-8 on 2018 Oct 20
|
Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X 5:100045 (2021)
Zhang CM, Imoto Y, Hikima T, Inoue T
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
UniProt ID: P04958 (564-864) Tetanus toxin
|
|
|
|
| Sample: |
Tetanus toxin monomer, 35 kDa Clostridium tetani protein
|
| Buffer: |
10 mM HEPES 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL45XU, SPring-8 on 2018 Oct 20
|
Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X 5:100045 (2021)
Zhang CM, Imoto Y, Hikima T, Inoue T
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
UniProt ID: P36056 (17-628) Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22, mitochondrial
|
|
|

|
| Sample: |
Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22, mitochondrial monomer, 70 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 May 1
|
Expression and analysis of the SAM-dependent RNA methyltransferase Rsm22 from Saccharomyces cerevisiae
Acta Crystallographica Section D Structural Biology 77(6) (2021)
Alam J, Rahman F, Sah-Teli S, Venkatesan R, Koski M, Autio K, Hiltunen J, Kastaniotis A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
UniProt ID: P36056 (17-628) dimeric Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22, mitochondrial
|
|
|

|
| Sample: |
Dimeric Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22, mitochondrial dimer, 141 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 May 1
|
Expression and analysis of the SAM-dependent RNA methyltransferase Rsm22 from Saccharomyces cerevisiae
Acta Crystallographica Section D Structural Biology 77(6) (2021)
Alam J, Rahman F, Sah-Teli S, Venkatesan R, Koski M, Autio K, Hiltunen J, Kastaniotis A
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.2 |
nm |
| VolumePorod |
516 |
nm3 |
|
|
UniProt ID: P05121 (24-402) Plasminogen activator inhibitor 1
UniProt ID: None (None-None) VHH-s-a93 (Ig module Nb93)
|
|
|
|
| Sample: |
Plasminogen activator inhibitor 1 monomer, 43 kDa Homo sapiens protein
VHH-s-a93 (Ig module Nb93) monomer, 13 kDa Vicugna pacos protein
|
| Buffer: |
30 mM BIS-TRIS pH 5.5, 300 mM sodium chloride, 5% v/v glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 5
|
Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity
International Journal of Molecular Sciences 21(16):5859 (2020)
Sillen M, Weeks S, Strelkov S, Declerck P
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
80 |
nm3 |
|
|