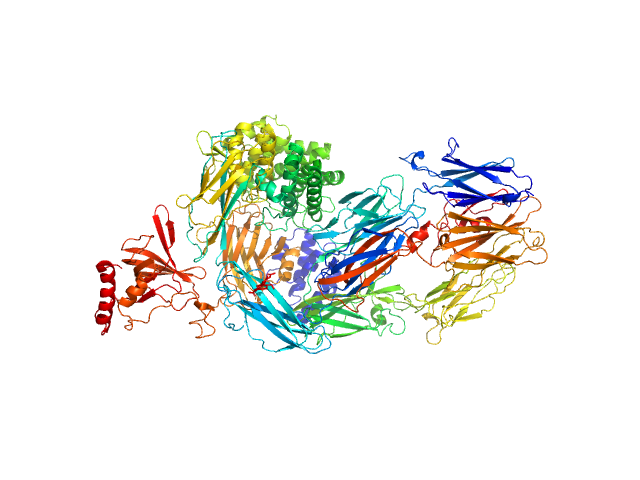
UniProt ID: P01031 (19-1676) Complement C5
|
|
|
|
| Sample: |
Complement C5 monomer, 186 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris pH, 75mM NaCl, and 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 12
|
The allosteric modulation of Complement C5 by knob domain peptides.
Elife 10 (2021)
Macpherson A, Laabei M, Ahdash Z, Graewert MA, Birtley JR, Schulze ME, Crennell S, Robinson SA, Holmes B, Oleinikovas V, Nilsson PH, Snowden J, Ellis V, Mollnes TE, Deane CM, Svergun D, Lawson AD, van den Elsen JM
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
392 |
nm3 |
|
|
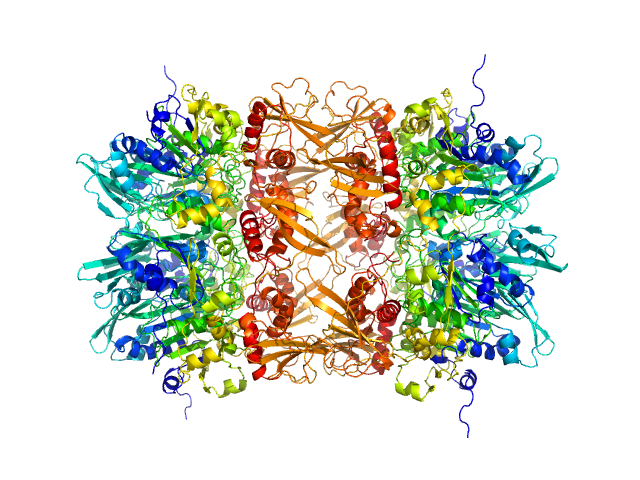
UniProt ID: B1ABI1 (None-None) gp11 encapsidation protein
|
|
|
|
| Sample: |
Gp11 encapsidation protein decamer, 446 kDa Lactococcus phage Phi28 protein
|
| Buffer: |
100 mM NaCl, 25 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2015 May 6
|
Lactococcus Phage ASCC Phi28 gp11
Mark White
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.3 |
nm |
|
|
UniProt ID: P04070 (43-461) Vitamin K-dependent protein C
|
|
|

|
| Sample: |
Vitamin K-dependent protein C monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 145 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 14
|
Zymogen and activated protein C have similar structural architecture.
J Biol Chem (2020)
Stojanovski BM, Pelc LA, Zuo X, Di Cera E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
|
|
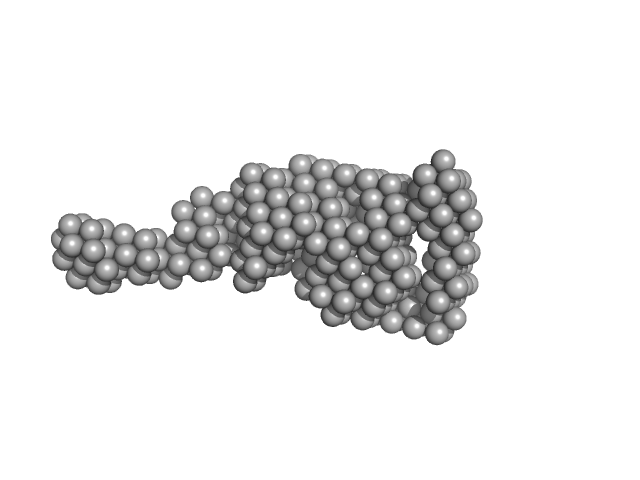
UniProt ID: P04070 (43-461) Vitamin K-dependent protein C
|
|
|
|
| Sample: |
Vitamin K-dependent protein C monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 145 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Aug 17
|
Zymogen and activated protein C have similar structural architecture.
J Biol Chem (2020)
Stojanovski BM, Pelc LA, Zuo X, Di Cera E
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
|
|
UniProt ID: Q9NP98 (1-174) N-ter construct of FATZ-1 (alias myozenin-1 or calsarcin-2)
|
|
|
|
| Sample: |
N-ter construct of FATZ-1 (alias myozenin-1 or calsarcin-2) monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 29
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: Q9NP98 (92-299) Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2)
|
|
|
|
| Sample: |
Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 18
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
3.9 |
nm |
| Dmax |
17.3 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
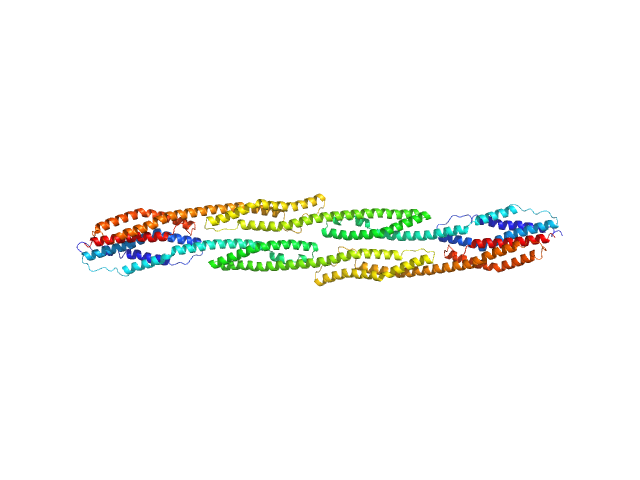
UniProt ID: P35609 (274-746) Rod domain of α-actinin-2
|
|
|
|
| Sample: |
Rod domain of α-actinin-2 dimer, 112 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 5
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
6.7 |
nm |
| Dmax |
27.2 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
UniProt ID: P35609 (1-894) Half dimer of α-actinin-2
|
|
|
|
| Sample: |
Half dimer of α-actinin-2 monomer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 18
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
5.3 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
172 |
nm3 |
|
|
UniProt ID: P35609 (274-746) Rod domain of α-actinin-2
UniProt ID: Q9NP98 (92-299) Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2)
|
|
|
|
| Sample: |
Rod domain of α-actinin-2 dimer, 112 kDa Homo sapiens protein
Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2) dimer, 43 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 18
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
7.6 |
nm |
| Dmax |
28.4 |
nm |
| VolumePorod |
371 |
nm3 |
|
|
UniProt ID: Q9NP98 (92-299) Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2)
UniProt ID: P35609 (1-894) Half dimer of α-actinin-2
|
|
|
|
| Sample: |
Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2) monomer, 22 kDa Homo sapiens protein
Half dimer of α-actinin-2 monomer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 18
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
5.8 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
242 |
nm3 |
|
|