UniProt ID: P11206 (None-None) Matrix protein
|
|
|
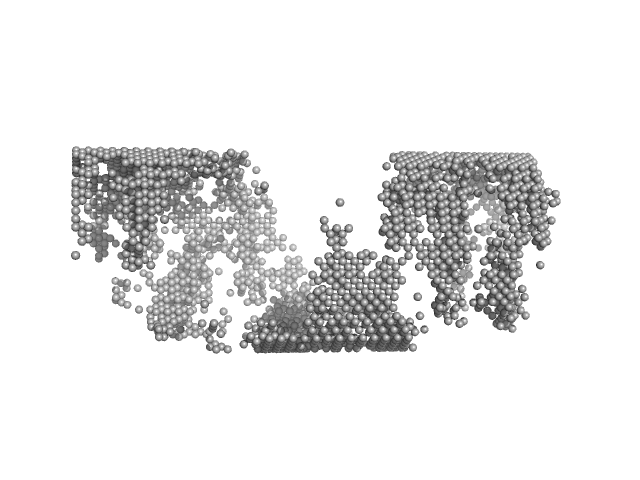
|
| Sample: |
Matrix protein, 40 kDa Newcastle disease virus … protein
|
| Buffer: |
STE buffer 100 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 11
|
Solution Structure, Self-Assembly, and Membrane Interactions of the Matrix Protein from Newcastle Disease Virus at Neutral and Acidic pH
Journal of Virology 93(6) (2019)
Shtykova E, Petoukhov M, Dadinova L, Fedorova N, Tashkin V, Timofeeva T, Ksenofontov A, Loshkarev N, Baratova L, Jeffries C, Svergun D, Batishchev O, García-Sastre A
|
|
|
UniProt ID: P11206 (None-None) Matrix protein
|
|
|
|
| Sample: |
Matrix protein dimer, 79 kDa Newcastle disease virus … protein
|
| Buffer: |
STE buffer 100 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 11
|
Solution Structure, Self-Assembly, and Membrane Interactions of the Matrix Protein from Newcastle Disease Virus at Neutral and Acidic pH
Journal of Virology 93(6) (2019)
Shtykova E, Petoukhov M, Dadinova L, Fedorova N, Tashkin V, Timofeeva T, Ksenofontov A, Loshkarev N, Baratova L, Jeffries C, Svergun D, Batishchev O, García-Sastre A
|
|
|
UniProt ID: Q5JRX3 (33-1036) Presequence protease, mitochondrial
|
|
|
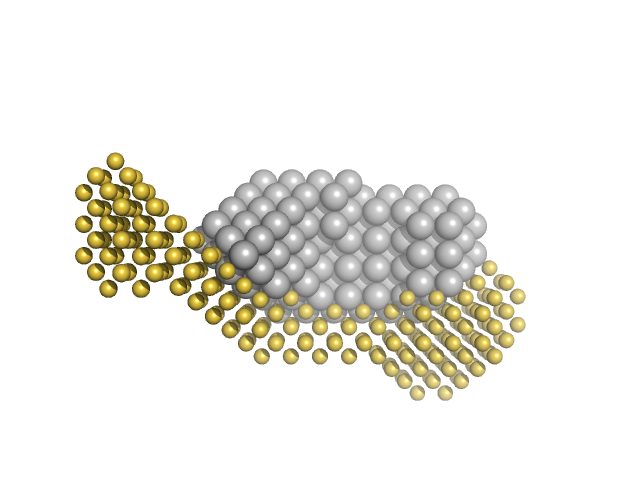
|
| Sample: |
Presequence protease, mitochondrial monomer, 115 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 4
|
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications 13(1) (2022)
Liang W, Wijaya J, Wei H, Noble A, Mancl J, Mo S, Lee D, Lin King J, Pan M, Liu C, Koehler C, Zhao M, Potter C, Carragher B, Li S, Tang W
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
UniProt ID: Q5JRX3 (33-1036) Presequence protease, mitochondrial
|
|
|
|
| Sample: |
Presequence protease, mitochondrial monomer, 115 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 7
|
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications 13(1) (2022)
Liang W, Wijaya J, Wei H, Noble A, Mancl J, Mo S, Lee D, Lin King J, Pan M, Liu C, Koehler C, Zhao M, Potter C, Carragher B, Li S, Tang W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
UniProt ID: Q5JRX3 (33-1036) Presequence protease, mitochondrial
UniProt ID: O75390 (1-27) Citrate synthase, mitochondrial
|
|
|
|
| Sample: |
Presequence protease, mitochondrial monomer, 115 kDa Homo sapiens protein
Citrate synthase, mitochondrial monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl , 20 mM EDTA, pH: 7.7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 4
|
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications 13(1) (2022)
Liang W, Wijaya J, Wei H, Noble A, Mancl J, Mo S, Lee D, Lin King J, Pan M, Liu C, Koehler C, Zhao M, Potter C, Carragher B, Li S, Tang W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
UniProt ID: Q5JRX3 (33-1036) Presequence protease, mitochondrial
UniProt ID: P05067 (688-711) Amyloid-beta precursor protein
|
|
|
|
| Sample: |
Presequence protease, mitochondrial monomer, 115 kDa Homo sapiens protein
Amyloid-beta precursor protein monomer, 4 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl , 20 mM EDTA, pH: 7.7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 7
|
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications 13(1) (2022)
Liang W, Wijaya J, Wei H, Noble A, Mancl J, Mo S, Lee D, Lin King J, Pan M, Liu C, Koehler C, Zhao M, Potter C, Carragher B, Li S, Tang W
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
UniProt ID: None (53-551) Alpha domain of Ag43a
|
|
|
|
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 3
|
Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms
npj Biofilms and Microbiomes 8(1) (2022)
Vo J, Ortiz G, Totsika M, Lo A, Hancock S, Whitten A, Hor L, Peters K, Ageorges V, Caccia N, Desvaux M, Schembri M, Paxman J, Heras B
|
| RgGuinier |
4.6 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
UniProt ID: P0DP33 (2-149) Calmodulin-1
UniProt ID: None (None-None) calcium ions
UniProt ID: P12497 (1-133) Gag-Pol polyprotein
|
|
|
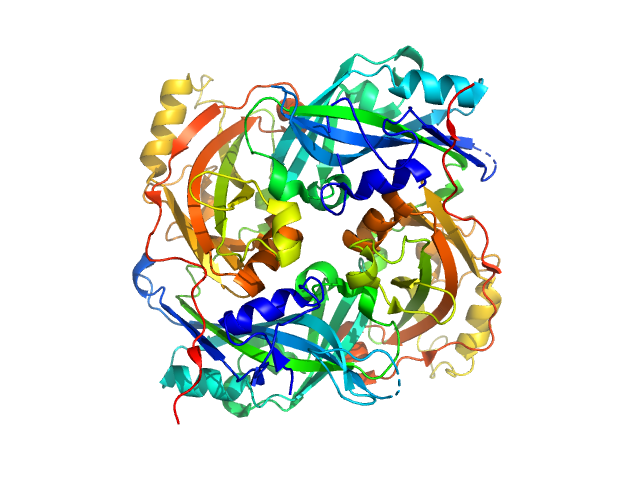
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Xenopus laevis protein
Calcium ions tetramer, 0 kDa
Gag-Pol polyprotein monomer, 15 kDa Human immunodeficiency virus … protein
|
| Buffer: |
50 mM MOPS, 5 mM CaCl2, 2 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, Australian Nuclear Science and Technology Organisation on 2010 Jun 23
|
Calmodulin binds a highly extended HIV-1 MA protein that refolds upon its release.
Biophys J 103(3):541-549 (2012)
Taylor JE, Chow JYH, Jeffries CM, Kwan AH, Duff AP, Hamilton WA, Trewhella J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
UniProt ID: P14616 (28-918) Insulin receptor-related protein
|
|
|
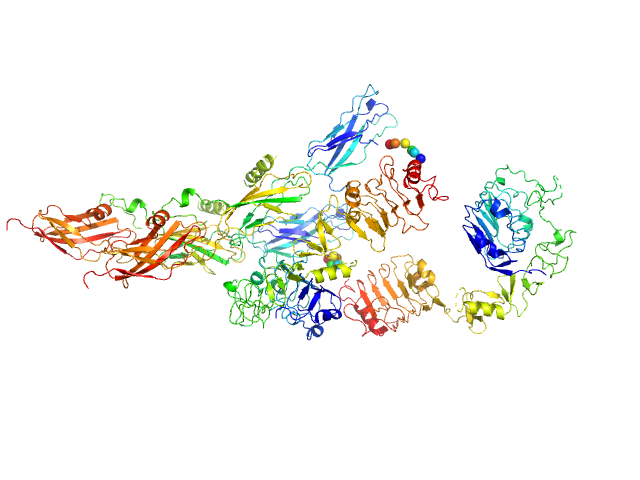
|
| Sample: |
Insulin receptor-related protein dimer, 201 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 22
|
The dimeric ectodomain of the alkali-sensing insulin receptor-related receptor (ectoIRR) has a droplike shape.
J Biol Chem 294(47):17790-17798 (2019)
Shtykova EV, Petoukhov MV, Mozhaev AA, Deyev IE, Dadinova LA, Loshkarev NA, Goryashchenko AS, Bocharov EV, Jeffries CM, Svergun DI, Batishchev OV, Petrenko AG
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
444 |
nm3 |
|
|
UniProt ID: P14616 (28-918) Insulin receptor-related protein
|
|
|
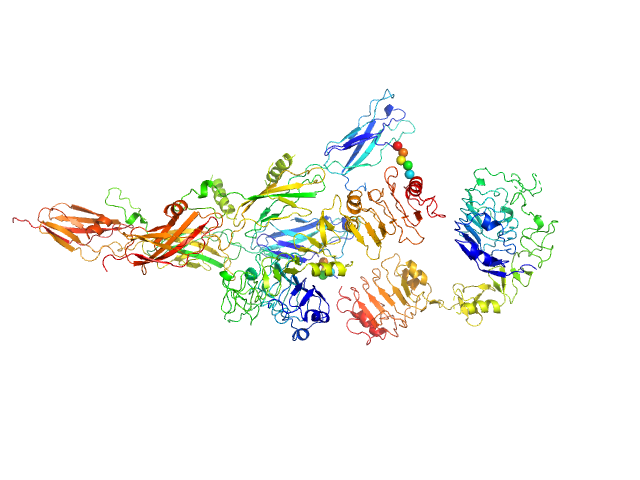
|
| Sample: |
Insulin receptor-related protein dimer, 201 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 22
|
The dimeric ectodomain of the alkali-sensing insulin receptor-related receptor (ectoIRR) has a droplike shape.
J Biol Chem 294(47):17790-17798 (2019)
Shtykova EV, Petoukhov MV, Mozhaev AA, Deyev IE, Dadinova LA, Loshkarev NA, Goryashchenko AS, Bocharov EV, Jeffries CM, Svergun DI, Batishchev OV, Petrenko AG
|
| RgGuinier |
5.3 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
430 |
nm3 |
|
|